
Anaeromyxobacter dehalogenans (strain 2CP-C)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Anaeromyxobacteraceae; Anaeromyxobacter; Anaeromyxobacter dehalogenans
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
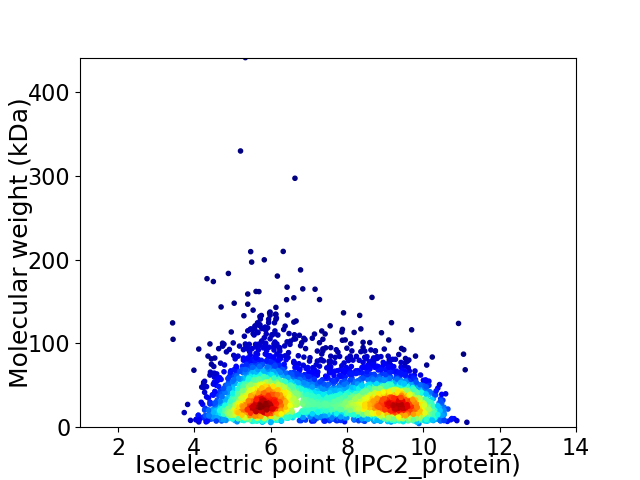
Virtual 2D-PAGE plot for 4345 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2IMM5|Q2IMM5_ANADE DNA-(apurinic or apyrimidinic site) lyase OS=Anaeromyxobacter dehalogenans (strain 2CP-C) OX=290397 GN=Adeh_0277 PE=3 SV=1
MM1 pKa = 7.14KK2 pKa = 9.79TLTKK6 pKa = 10.56LALTLIAVAGLQACDD21 pKa = 3.94PYY23 pKa = 11.24EE24 pKa = 4.48DD25 pKa = 4.14EE26 pKa = 4.65NKK28 pKa = 10.45GVPSVINVISSDD40 pKa = 3.09SGTAYY45 pKa = 10.28EE46 pKa = 4.47GTNTGTSWTISTPSAFVLFVKK67 pKa = 10.5VNAGKK72 pKa = 10.43LLDD75 pKa = 4.23GASIQATTDD84 pKa = 3.02DD85 pKa = 4.49CTPANDD91 pKa = 3.64WLSVNGAGPGNWMSCYY107 pKa = 10.06YY108 pKa = 8.81PQSPAGFEE116 pKa = 4.14GASVVLYY123 pKa = 8.04PGPSISTVDD132 pKa = 2.97SWFSLSEE139 pKa = 4.04LEE141 pKa = 4.49PGIFNLAGTVSDD153 pKa = 4.01KK154 pKa = 10.88QGHH157 pKa = 6.45PLAIDD162 pKa = 3.2VNAFVNDD169 pKa = 4.23LAAAQSDD176 pKa = 4.02TAGTEE181 pKa = 3.62ITTEE185 pKa = 3.97FSDD188 pKa = 4.6AGVNMTNVQLWEE200 pKa = 4.28APDD203 pKa = 3.46VSGAPGTWTAVATTTPYY220 pKa = 9.69VHH222 pKa = 7.05AGVAVGSTWWYY233 pKa = 10.83KK234 pKa = 8.59VTADD238 pKa = 3.88TNGFPATAASFEE250 pKa = 4.27TAPVSVTTEE259 pKa = 3.82PPAPP263 pKa = 3.85
MM1 pKa = 7.14KK2 pKa = 9.79TLTKK6 pKa = 10.56LALTLIAVAGLQACDD21 pKa = 3.94PYY23 pKa = 11.24EE24 pKa = 4.48DD25 pKa = 4.14EE26 pKa = 4.65NKK28 pKa = 10.45GVPSVINVISSDD40 pKa = 3.09SGTAYY45 pKa = 10.28EE46 pKa = 4.47GTNTGTSWTISTPSAFVLFVKK67 pKa = 10.5VNAGKK72 pKa = 10.43LLDD75 pKa = 4.23GASIQATTDD84 pKa = 3.02DD85 pKa = 4.49CTPANDD91 pKa = 3.64WLSVNGAGPGNWMSCYY107 pKa = 10.06YY108 pKa = 8.81PQSPAGFEE116 pKa = 4.14GASVVLYY123 pKa = 8.04PGPSISTVDD132 pKa = 2.97SWFSLSEE139 pKa = 4.04LEE141 pKa = 4.49PGIFNLAGTVSDD153 pKa = 4.01KK154 pKa = 10.88QGHH157 pKa = 6.45PLAIDD162 pKa = 3.2VNAFVNDD169 pKa = 4.23LAAAQSDD176 pKa = 4.02TAGTEE181 pKa = 3.62ITTEE185 pKa = 3.97FSDD188 pKa = 4.6AGVNMTNVQLWEE200 pKa = 4.28APDD203 pKa = 3.46VSGAPGTWTAVATTTPYY220 pKa = 9.69VHH222 pKa = 7.05AGVAVGSTWWYY233 pKa = 10.83KK234 pKa = 8.59VTADD238 pKa = 3.88TNGFPATAASFEE250 pKa = 4.27TAPVSVTTEE259 pKa = 3.82PPAPP263 pKa = 3.85
Molecular weight: 27.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2IIL5|Q2IIL5_ANADE Molybdopterin binding domain protein OS=Anaeromyxobacter dehalogenans (strain 2CP-C) OX=290397 GN=Adeh_1720 PE=4 SV=1
MM1 pKa = 7.76APRR4 pKa = 11.84APLPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84SPGCPRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GPRR28 pKa = 11.84LPRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84SRR36 pKa = 11.84LAGAGAARR44 pKa = 11.84RR45 pKa = 11.84GGPRR49 pKa = 11.84GGRR52 pKa = 11.84ARR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84PDD63 pKa = 2.62GRR65 pKa = 11.84AARR68 pKa = 11.84RR69 pKa = 11.84QRR71 pKa = 11.84RR72 pKa = 11.84GAPGGRR78 pKa = 11.84RR79 pKa = 11.84GVRR82 pKa = 11.84GARR85 pKa = 11.84RR86 pKa = 11.84GRR88 pKa = 11.84GGRR91 pKa = 11.84GAGGAPGGSGRR102 pKa = 11.84DD103 pKa = 3.28GGGRR107 pKa = 11.84GRR109 pKa = 11.84ARR111 pKa = 11.84ARR113 pKa = 11.84RR114 pKa = 11.84HH115 pKa = 5.75AGPAAGARR123 pKa = 11.84RR124 pKa = 11.84AARR127 pKa = 11.84GLRR130 pKa = 11.84GRR132 pKa = 11.84LARR135 pKa = 11.84RR136 pKa = 11.84ARR138 pKa = 11.84RR139 pKa = 11.84HH140 pKa = 4.64RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84ARR145 pKa = 11.84RR146 pKa = 11.84GGRR149 pKa = 11.84LAARR153 pKa = 11.84GGRR156 pKa = 11.84AGGARR161 pKa = 11.84ALGAQPGGPGRR172 pKa = 11.84PRR174 pKa = 11.84GGSRR178 pKa = 11.84GRR180 pKa = 11.84GGGDD184 pKa = 2.63RR185 pKa = 11.84RR186 pKa = 11.84EE187 pKa = 3.93RR188 pKa = 11.84PGRR191 pKa = 11.84GGGAARR197 pKa = 11.84AAARR201 pKa = 11.84PGAQPGPRR209 pKa = 11.84ARR211 pKa = 11.84RR212 pKa = 11.84GRR214 pKa = 11.84GARR217 pKa = 11.84AGRR220 pKa = 11.84GGGLGGAAGARR231 pKa = 11.84GAAALAGSGADD242 pKa = 2.83RR243 pKa = 11.84GRR245 pKa = 11.84RR246 pKa = 11.84GRR248 pKa = 11.84AGPSRR253 pKa = 11.84GSRR256 pKa = 11.84RR257 pKa = 11.84DD258 pKa = 2.96RR259 pKa = 11.84RR260 pKa = 11.84ARR262 pKa = 11.84GGGARR267 pKa = 11.84LVRR270 pKa = 11.84ADD272 pKa = 3.52RR273 pKa = 11.84GGGRR277 pKa = 11.84GRR279 pKa = 11.84ARR281 pKa = 11.84GAAGGARR288 pKa = 11.84GRR290 pKa = 11.84DD291 pKa = 3.22GALRR295 pKa = 11.84GGGAEE300 pKa = 3.76RR301 pKa = 11.84GSRR304 pKa = 11.84RR305 pKa = 11.84GGARR309 pKa = 11.84CARR312 pKa = 11.84GPAARR317 pKa = 11.84DD318 pKa = 3.46RR319 pKa = 11.84VPGAPVPGARR329 pKa = 11.84GPAGGRR335 pKa = 11.84GRR337 pKa = 11.84PGGARR342 pKa = 11.84GAGAHH347 pKa = 5.49GRR349 pKa = 11.84RR350 pKa = 11.84GGRR353 pKa = 11.84GRR355 pKa = 11.84GAPAPAPRR363 pKa = 11.84PRR365 pKa = 11.84PRR367 pKa = 11.84RR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84AARR373 pKa = 11.84RR374 pKa = 11.84RR375 pKa = 11.84RR376 pKa = 11.84ARR378 pKa = 11.84AGPARR383 pKa = 11.84RR384 pKa = 11.84PGLRR388 pKa = 11.84RR389 pKa = 11.84PGPRR393 pKa = 11.84PRR395 pKa = 11.84RR396 pKa = 11.84GAGRR400 pKa = 11.84APGGRR405 pKa = 11.84PGTAGRR411 pKa = 11.84VAAARR416 pKa = 11.84RR417 pKa = 11.84RR418 pKa = 11.84RR419 pKa = 11.84DD420 pKa = 2.99PCAARR425 pKa = 11.84RR426 pKa = 11.84DD427 pKa = 4.06APAHH431 pKa = 5.38RR432 pKa = 11.84RR433 pKa = 11.84RR434 pKa = 11.84RR435 pKa = 11.84RR436 pKa = 11.84ARR438 pKa = 11.84AGGAAGRR445 pKa = 11.84RR446 pKa = 11.84RR447 pKa = 11.84DD448 pKa = 3.39PARR451 pKa = 11.84AGRR454 pKa = 11.84GAAPARR460 pKa = 11.84GGARR464 pKa = 11.84CAPGGAHH471 pKa = 6.63RR472 pKa = 11.84RR473 pKa = 11.84DD474 pKa = 3.77RR475 pKa = 11.84RR476 pKa = 11.84PGGAGHH482 pKa = 7.12RR483 pKa = 11.84PGAAGRR489 pKa = 11.84GRR491 pKa = 11.84RR492 pKa = 11.84ARR494 pKa = 11.84GRR496 pKa = 11.84RR497 pKa = 11.84AGGAARR503 pKa = 11.84GRR505 pKa = 11.84RR506 pKa = 11.84GAGPGRR512 pKa = 11.84AALGAGAGARR522 pKa = 11.84GGAGGARR529 pKa = 11.84RR530 pKa = 11.84RR531 pKa = 11.84RR532 pKa = 11.84LALRR536 pKa = 11.84AGGGAGRR543 pKa = 11.84RR544 pKa = 11.84GRR546 pKa = 11.84GEE548 pKa = 3.87RR549 pKa = 11.84GGAAHH554 pKa = 7.44RR555 pKa = 11.84APRR558 pKa = 11.84RR559 pKa = 11.84RR560 pKa = 11.84VRR562 pKa = 11.84PGGGVRR568 pKa = 11.84ARR570 pKa = 11.84QDD572 pKa = 2.72RR573 pKa = 11.84RR574 pKa = 11.84AGRR577 pKa = 11.84GEE579 pKa = 3.88AAGGLAGAPGRR590 pKa = 11.84ARR592 pKa = 11.84GRR594 pKa = 11.84RR595 pKa = 11.84GDD597 pKa = 3.35RR598 pKa = 11.84GARR601 pKa = 11.84AARR604 pKa = 11.84RR605 pKa = 11.84PRR607 pKa = 11.84RR608 pKa = 11.84RR609 pKa = 11.84AGHH612 pKa = 6.02RR613 pKa = 11.84RR614 pKa = 11.84AAHH617 pKa = 5.68RR618 pKa = 11.84GSRR621 pKa = 11.84GGAGGGGAGHH631 pKa = 7.02RR632 pKa = 11.84PAPPRR637 pKa = 11.84GGRR640 pKa = 11.84AAAGGAAEE648 pKa = 4.12RR649 pKa = 11.84LPRR652 pKa = 11.84ARR654 pKa = 11.84AARR657 pKa = 11.84RR658 pKa = 11.84GGGAVAAPGGRR669 pKa = 11.84AADD672 pKa = 3.71AALKK676 pKa = 10.42AA677 pKa = 4.08
MM1 pKa = 7.76APRR4 pKa = 11.84APLPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84SPGCPRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GPRR28 pKa = 11.84LPRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84SRR36 pKa = 11.84LAGAGAARR44 pKa = 11.84RR45 pKa = 11.84GGPRR49 pKa = 11.84GGRR52 pKa = 11.84ARR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84PDD63 pKa = 2.62GRR65 pKa = 11.84AARR68 pKa = 11.84RR69 pKa = 11.84QRR71 pKa = 11.84RR72 pKa = 11.84GAPGGRR78 pKa = 11.84RR79 pKa = 11.84GVRR82 pKa = 11.84GARR85 pKa = 11.84RR86 pKa = 11.84GRR88 pKa = 11.84GGRR91 pKa = 11.84GAGGAPGGSGRR102 pKa = 11.84DD103 pKa = 3.28GGGRR107 pKa = 11.84GRR109 pKa = 11.84ARR111 pKa = 11.84ARR113 pKa = 11.84RR114 pKa = 11.84HH115 pKa = 5.75AGPAAGARR123 pKa = 11.84RR124 pKa = 11.84AARR127 pKa = 11.84GLRR130 pKa = 11.84GRR132 pKa = 11.84LARR135 pKa = 11.84RR136 pKa = 11.84ARR138 pKa = 11.84RR139 pKa = 11.84HH140 pKa = 4.64RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84ARR145 pKa = 11.84RR146 pKa = 11.84GGRR149 pKa = 11.84LAARR153 pKa = 11.84GGRR156 pKa = 11.84AGGARR161 pKa = 11.84ALGAQPGGPGRR172 pKa = 11.84PRR174 pKa = 11.84GGSRR178 pKa = 11.84GRR180 pKa = 11.84GGGDD184 pKa = 2.63RR185 pKa = 11.84RR186 pKa = 11.84EE187 pKa = 3.93RR188 pKa = 11.84PGRR191 pKa = 11.84GGGAARR197 pKa = 11.84AAARR201 pKa = 11.84PGAQPGPRR209 pKa = 11.84ARR211 pKa = 11.84RR212 pKa = 11.84GRR214 pKa = 11.84GARR217 pKa = 11.84AGRR220 pKa = 11.84GGGLGGAAGARR231 pKa = 11.84GAAALAGSGADD242 pKa = 2.83RR243 pKa = 11.84GRR245 pKa = 11.84RR246 pKa = 11.84GRR248 pKa = 11.84AGPSRR253 pKa = 11.84GSRR256 pKa = 11.84RR257 pKa = 11.84DD258 pKa = 2.96RR259 pKa = 11.84RR260 pKa = 11.84ARR262 pKa = 11.84GGGARR267 pKa = 11.84LVRR270 pKa = 11.84ADD272 pKa = 3.52RR273 pKa = 11.84GGGRR277 pKa = 11.84GRR279 pKa = 11.84ARR281 pKa = 11.84GAAGGARR288 pKa = 11.84GRR290 pKa = 11.84DD291 pKa = 3.22GALRR295 pKa = 11.84GGGAEE300 pKa = 3.76RR301 pKa = 11.84GSRR304 pKa = 11.84RR305 pKa = 11.84GGARR309 pKa = 11.84CARR312 pKa = 11.84GPAARR317 pKa = 11.84DD318 pKa = 3.46RR319 pKa = 11.84VPGAPVPGARR329 pKa = 11.84GPAGGRR335 pKa = 11.84GRR337 pKa = 11.84PGGARR342 pKa = 11.84GAGAHH347 pKa = 5.49GRR349 pKa = 11.84RR350 pKa = 11.84GGRR353 pKa = 11.84GRR355 pKa = 11.84GAPAPAPRR363 pKa = 11.84PRR365 pKa = 11.84PRR367 pKa = 11.84RR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84AARR373 pKa = 11.84RR374 pKa = 11.84RR375 pKa = 11.84RR376 pKa = 11.84ARR378 pKa = 11.84AGPARR383 pKa = 11.84RR384 pKa = 11.84PGLRR388 pKa = 11.84RR389 pKa = 11.84PGPRR393 pKa = 11.84PRR395 pKa = 11.84RR396 pKa = 11.84GAGRR400 pKa = 11.84APGGRR405 pKa = 11.84PGTAGRR411 pKa = 11.84VAAARR416 pKa = 11.84RR417 pKa = 11.84RR418 pKa = 11.84RR419 pKa = 11.84DD420 pKa = 2.99PCAARR425 pKa = 11.84RR426 pKa = 11.84DD427 pKa = 4.06APAHH431 pKa = 5.38RR432 pKa = 11.84RR433 pKa = 11.84RR434 pKa = 11.84RR435 pKa = 11.84RR436 pKa = 11.84ARR438 pKa = 11.84AGGAAGRR445 pKa = 11.84RR446 pKa = 11.84RR447 pKa = 11.84DD448 pKa = 3.39PARR451 pKa = 11.84AGRR454 pKa = 11.84GAAPARR460 pKa = 11.84GGARR464 pKa = 11.84CAPGGAHH471 pKa = 6.63RR472 pKa = 11.84RR473 pKa = 11.84DD474 pKa = 3.77RR475 pKa = 11.84RR476 pKa = 11.84PGGAGHH482 pKa = 7.12RR483 pKa = 11.84PGAAGRR489 pKa = 11.84GRR491 pKa = 11.84RR492 pKa = 11.84ARR494 pKa = 11.84GRR496 pKa = 11.84RR497 pKa = 11.84AGGAARR503 pKa = 11.84GRR505 pKa = 11.84RR506 pKa = 11.84GAGPGRR512 pKa = 11.84AALGAGAGARR522 pKa = 11.84GGAGGARR529 pKa = 11.84RR530 pKa = 11.84RR531 pKa = 11.84RR532 pKa = 11.84LALRR536 pKa = 11.84AGGGAGRR543 pKa = 11.84RR544 pKa = 11.84GRR546 pKa = 11.84GEE548 pKa = 3.87RR549 pKa = 11.84GGAAHH554 pKa = 7.44RR555 pKa = 11.84APRR558 pKa = 11.84RR559 pKa = 11.84RR560 pKa = 11.84VRR562 pKa = 11.84PGGGVRR568 pKa = 11.84ARR570 pKa = 11.84QDD572 pKa = 2.72RR573 pKa = 11.84RR574 pKa = 11.84AGRR577 pKa = 11.84GEE579 pKa = 3.88AAGGLAGAPGRR590 pKa = 11.84ARR592 pKa = 11.84GRR594 pKa = 11.84RR595 pKa = 11.84GDD597 pKa = 3.35RR598 pKa = 11.84GARR601 pKa = 11.84AARR604 pKa = 11.84RR605 pKa = 11.84PRR607 pKa = 11.84RR608 pKa = 11.84RR609 pKa = 11.84AGHH612 pKa = 6.02RR613 pKa = 11.84RR614 pKa = 11.84AAHH617 pKa = 5.68RR618 pKa = 11.84GSRR621 pKa = 11.84GGAGGGGAGHH631 pKa = 7.02RR632 pKa = 11.84PAPPRR637 pKa = 11.84GGRR640 pKa = 11.84AAAGGAAEE648 pKa = 4.12RR649 pKa = 11.84LPRR652 pKa = 11.84ARR654 pKa = 11.84AARR657 pKa = 11.84RR658 pKa = 11.84GGGAVAAPGGRR669 pKa = 11.84AADD672 pKa = 3.71AALKK676 pKa = 10.42AA677 pKa = 4.08
Molecular weight: 68.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1516487 |
38 |
4074 |
349.0 |
37.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.967 ± 0.084 | 0.884 ± 0.02 |
5.127 ± 0.024 | 6.159 ± 0.038 |
2.962 ± 0.023 | 9.348 ± 0.044 |
1.963 ± 0.017 | 2.933 ± 0.025 |
2.338 ± 0.036 | 10.854 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.635 ± 0.016 | 1.526 ± 0.019 |
6.364 ± 0.038 | 2.476 ± 0.019 |
9.386 ± 0.053 | 4.21 ± 0.024 |
4.387 ± 0.029 | 8.371 ± 0.034 |
1.284 ± 0.014 | 1.824 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
