
Cochliobolus heterostrophus (strain C5 / ATCC 48332 / race O) (Southern corn leaf blight fungus) (Bipolaris maydis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Pleosporineae; Pleosporaceae; Bipolaris; Bipolaris maydis
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
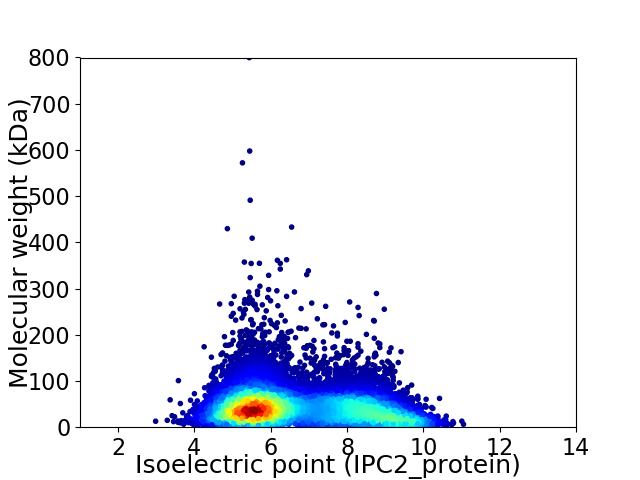
Virtual 2D-PAGE plot for 12857 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M2TGX7|M2TGX7_COCH5 Uncharacterized protein OS=Cochliobolus heterostrophus (strain C5 / ATCC 48332 / race O) OX=701091 GN=COCHEDRAFT_1024357 PE=4 SV=1
MM1 pKa = 6.64TAAVSTNGTPRR12 pKa = 11.84YY13 pKa = 9.49RR14 pKa = 11.84STLSSPSTEE23 pKa = 4.18SVTLASPLSPVPVSDD38 pKa = 4.33FEE40 pKa = 6.48DD41 pKa = 3.73IFTMSPIDD49 pKa = 3.74SEE51 pKa = 4.82TEE53 pKa = 3.54IEE55 pKa = 4.25TDD57 pKa = 3.36SNAAPVPMSFEE68 pKa = 4.26VPLGPLSGYY77 pKa = 10.41GGAPKK82 pKa = 9.88PVPKK86 pKa = 10.07YY87 pKa = 10.02KK88 pKa = 10.6KK89 pKa = 9.49PATRR93 pKa = 11.84LIKK96 pKa = 10.07PVSEE100 pKa = 4.19QPDD103 pKa = 3.6YY104 pKa = 11.21VYY106 pKa = 11.2SFKK109 pKa = 11.08GYY111 pKa = 10.36DD112 pKa = 3.26ADD114 pKa = 4.95ALVLTDD120 pKa = 3.54QQINSLGQAVDD131 pKa = 4.23GKK133 pKa = 10.3QHH135 pKa = 6.68DD136 pKa = 4.69AEE138 pKa = 4.56PMGQSSPLYY147 pKa = 10.83GEE149 pKa = 4.32TDD151 pKa = 3.41SDD153 pKa = 4.18DD154 pKa = 4.34SDD156 pKa = 3.95PFIYY160 pKa = 10.46DD161 pKa = 3.89HH162 pKa = 7.27IILPSSSSSSSSDD175 pKa = 3.3YY176 pKa = 11.46YY177 pKa = 11.45NAVDD181 pKa = 4.06DD182 pKa = 4.53DD183 pKa = 4.29QYY185 pKa = 11.67NASDD189 pKa = 4.14EE190 pKa = 4.38GDD192 pKa = 3.69SPSDD196 pKa = 3.4NNSFEE201 pKa = 4.8DD202 pKa = 3.91EE203 pKa = 4.36SAFNSNSDD211 pKa = 3.45DD212 pKa = 4.18DD213 pKa = 4.92AFEE216 pKa = 4.48AHH218 pKa = 6.79DD219 pKa = 4.34TNNDD223 pKa = 3.76DD224 pKa = 4.47VGSEE228 pKa = 4.19PDD230 pKa = 3.68SKK232 pKa = 10.95PPSSPSSAASGLYY245 pKa = 10.61FNDD248 pKa = 3.76EE249 pKa = 4.28ADD251 pKa = 3.85SEE253 pKa = 4.76HH254 pKa = 7.09NNDD257 pKa = 5.17DD258 pKa = 4.92DD259 pKa = 5.96KK260 pKa = 11.95DD261 pKa = 5.0SNDD264 pKa = 4.99SNDD267 pKa = 5.19DD268 pKa = 4.11DD269 pKa = 6.78DD270 pKa = 6.02NDD272 pKa = 4.63SNNDD276 pKa = 3.58NDD278 pKa = 6.14DD279 pKa = 4.6DD280 pKa = 6.13DD281 pKa = 5.55NDD283 pKa = 4.42PNRR286 pKa = 11.84SDD288 pKa = 4.16NDD290 pKa = 3.28NDD292 pKa = 3.42EE293 pKa = 5.03HH294 pKa = 8.91KK295 pKa = 11.04DD296 pKa = 3.79EE297 pKa = 4.39EE298 pKa = 4.65DD299 pKa = 3.22EE300 pKa = 4.9RR301 pKa = 11.84NNDD304 pKa = 3.33DD305 pKa = 3.72YY306 pKa = 11.88YY307 pKa = 11.65NPEE310 pKa = 4.82DD311 pKa = 4.56YY312 pKa = 11.09FIDD315 pKa = 3.78TNNYY319 pKa = 9.71NYY321 pKa = 10.83SDD323 pKa = 4.29DD324 pKa = 4.36NEE326 pKa = 4.98SGDD329 pKa = 3.87SSSRR333 pKa = 11.84CSNLDD338 pKa = 3.07SHH340 pKa = 7.06PLYY343 pKa = 10.91LPAPIYY349 pKa = 10.76AIAPYY354 pKa = 8.75LTYY357 pKa = 10.34QSPPTAPTMLPPVPHH372 pKa = 6.24NTPLALPSNAPSTDD386 pKa = 2.82PRR388 pKa = 11.84YY389 pKa = 10.43SDD391 pKa = 4.54LGTLEE396 pKa = 4.0ILHH399 pKa = 5.4PHH401 pKa = 5.97IRR403 pKa = 11.84NMIYY407 pKa = 10.7GFVLADD413 pKa = 3.61SPTATSEE420 pKa = 4.09EE421 pKa = 4.49NNTTGAQSDD430 pKa = 3.32IGVRR434 pKa = 11.84HH435 pKa = 5.15STAMADD441 pKa = 3.96RR442 pKa = 11.84NNMLLVCWAMRR453 pKa = 11.84EE454 pKa = 4.02EE455 pKa = 4.32VLDD458 pKa = 4.37FLALPFSTSGDD469 pKa = 3.6EE470 pKa = 3.97
MM1 pKa = 6.64TAAVSTNGTPRR12 pKa = 11.84YY13 pKa = 9.49RR14 pKa = 11.84STLSSPSTEE23 pKa = 4.18SVTLASPLSPVPVSDD38 pKa = 4.33FEE40 pKa = 6.48DD41 pKa = 3.73IFTMSPIDD49 pKa = 3.74SEE51 pKa = 4.82TEE53 pKa = 3.54IEE55 pKa = 4.25TDD57 pKa = 3.36SNAAPVPMSFEE68 pKa = 4.26VPLGPLSGYY77 pKa = 10.41GGAPKK82 pKa = 9.88PVPKK86 pKa = 10.07YY87 pKa = 10.02KK88 pKa = 10.6KK89 pKa = 9.49PATRR93 pKa = 11.84LIKK96 pKa = 10.07PVSEE100 pKa = 4.19QPDD103 pKa = 3.6YY104 pKa = 11.21VYY106 pKa = 11.2SFKK109 pKa = 11.08GYY111 pKa = 10.36DD112 pKa = 3.26ADD114 pKa = 4.95ALVLTDD120 pKa = 3.54QQINSLGQAVDD131 pKa = 4.23GKK133 pKa = 10.3QHH135 pKa = 6.68DD136 pKa = 4.69AEE138 pKa = 4.56PMGQSSPLYY147 pKa = 10.83GEE149 pKa = 4.32TDD151 pKa = 3.41SDD153 pKa = 4.18DD154 pKa = 4.34SDD156 pKa = 3.95PFIYY160 pKa = 10.46DD161 pKa = 3.89HH162 pKa = 7.27IILPSSSSSSSSDD175 pKa = 3.3YY176 pKa = 11.46YY177 pKa = 11.45NAVDD181 pKa = 4.06DD182 pKa = 4.53DD183 pKa = 4.29QYY185 pKa = 11.67NASDD189 pKa = 4.14EE190 pKa = 4.38GDD192 pKa = 3.69SPSDD196 pKa = 3.4NNSFEE201 pKa = 4.8DD202 pKa = 3.91EE203 pKa = 4.36SAFNSNSDD211 pKa = 3.45DD212 pKa = 4.18DD213 pKa = 4.92AFEE216 pKa = 4.48AHH218 pKa = 6.79DD219 pKa = 4.34TNNDD223 pKa = 3.76DD224 pKa = 4.47VGSEE228 pKa = 4.19PDD230 pKa = 3.68SKK232 pKa = 10.95PPSSPSSAASGLYY245 pKa = 10.61FNDD248 pKa = 3.76EE249 pKa = 4.28ADD251 pKa = 3.85SEE253 pKa = 4.76HH254 pKa = 7.09NNDD257 pKa = 5.17DD258 pKa = 4.92DD259 pKa = 5.96KK260 pKa = 11.95DD261 pKa = 5.0SNDD264 pKa = 4.99SNDD267 pKa = 5.19DD268 pKa = 4.11DD269 pKa = 6.78DD270 pKa = 6.02NDD272 pKa = 4.63SNNDD276 pKa = 3.58NDD278 pKa = 6.14DD279 pKa = 4.6DD280 pKa = 6.13DD281 pKa = 5.55NDD283 pKa = 4.42PNRR286 pKa = 11.84SDD288 pKa = 4.16NDD290 pKa = 3.28NDD292 pKa = 3.42EE293 pKa = 5.03HH294 pKa = 8.91KK295 pKa = 11.04DD296 pKa = 3.79EE297 pKa = 4.39EE298 pKa = 4.65DD299 pKa = 3.22EE300 pKa = 4.9RR301 pKa = 11.84NNDD304 pKa = 3.33DD305 pKa = 3.72YY306 pKa = 11.88YY307 pKa = 11.65NPEE310 pKa = 4.82DD311 pKa = 4.56YY312 pKa = 11.09FIDD315 pKa = 3.78TNNYY319 pKa = 9.71NYY321 pKa = 10.83SDD323 pKa = 4.29DD324 pKa = 4.36NEE326 pKa = 4.98SGDD329 pKa = 3.87SSSRR333 pKa = 11.84CSNLDD338 pKa = 3.07SHH340 pKa = 7.06PLYY343 pKa = 10.91LPAPIYY349 pKa = 10.76AIAPYY354 pKa = 8.75LTYY357 pKa = 10.34QSPPTAPTMLPPVPHH372 pKa = 6.24NTPLALPSNAPSTDD386 pKa = 2.82PRR388 pKa = 11.84YY389 pKa = 10.43SDD391 pKa = 4.54LGTLEE396 pKa = 4.0ILHH399 pKa = 5.4PHH401 pKa = 5.97IRR403 pKa = 11.84NMIYY407 pKa = 10.7GFVLADD413 pKa = 3.61SPTATSEE420 pKa = 4.09EE421 pKa = 4.49NNTTGAQSDD430 pKa = 3.32IGVRR434 pKa = 11.84HH435 pKa = 5.15STAMADD441 pKa = 3.96RR442 pKa = 11.84NNMLLVCWAMRR453 pKa = 11.84EE454 pKa = 4.02EE455 pKa = 4.32VLDD458 pKa = 4.37FLALPFSTSGDD469 pKa = 3.6EE470 pKa = 3.97
Molecular weight: 51.24 kDa
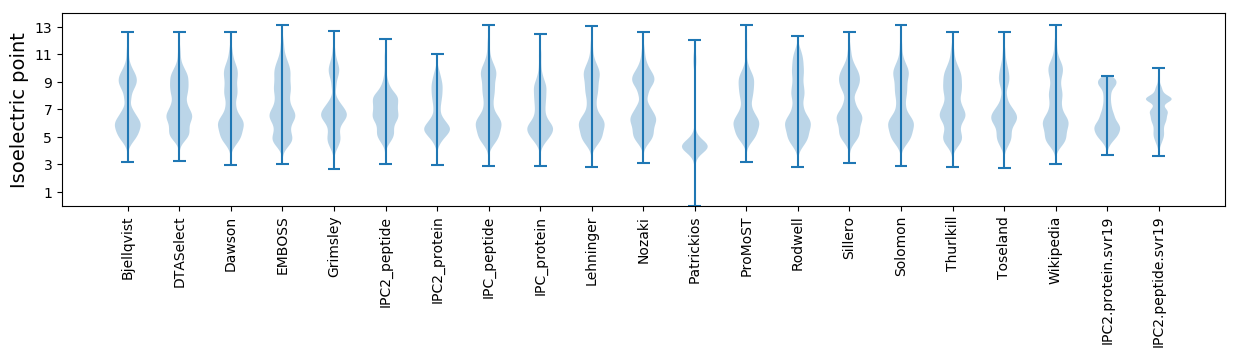
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M2TGL3|M2TGL3_COCH5 Uncharacterized protein OS=Cochliobolus heterostrophus (strain C5 / ATCC 48332 / race O) OX=701091 GN=COCHEDRAFT_1117819 PE=4 SV=1
MM1 pKa = 7.53PLARR5 pKa = 11.84RR6 pKa = 11.84SAPRR10 pKa = 11.84TARR13 pKa = 11.84TTRR16 pKa = 11.84TTARR20 pKa = 11.84PSLKK24 pKa = 9.13TRR26 pKa = 11.84LMGGGRR32 pKa = 11.84NTRR35 pKa = 11.84THH37 pKa = 6.06TKK39 pKa = 9.71RR40 pKa = 11.84APAGTTTTTTTTTRR54 pKa = 11.84TTRR57 pKa = 11.84TTGGRR62 pKa = 11.84HH63 pKa = 3.76GHH65 pKa = 6.51AAPVHH70 pKa = 4.63HH71 pKa = 7.34HH72 pKa = 6.34KK73 pKa = 10.63RR74 pKa = 11.84HH75 pKa = 4.81ATMGDD80 pKa = 3.45KK81 pKa = 10.91VSGMMMKK88 pKa = 10.48LRR90 pKa = 11.84GSLTRR95 pKa = 11.84RR96 pKa = 11.84PGLKK100 pKa = 10.1AAGTRR105 pKa = 11.84RR106 pKa = 11.84MHH108 pKa = 5.92GTDD111 pKa = 2.72GRR113 pKa = 11.84NARR116 pKa = 11.84RR117 pKa = 11.84VYY119 pKa = 10.89
MM1 pKa = 7.53PLARR5 pKa = 11.84RR6 pKa = 11.84SAPRR10 pKa = 11.84TARR13 pKa = 11.84TTRR16 pKa = 11.84TTARR20 pKa = 11.84PSLKK24 pKa = 9.13TRR26 pKa = 11.84LMGGGRR32 pKa = 11.84NTRR35 pKa = 11.84THH37 pKa = 6.06TKK39 pKa = 9.71RR40 pKa = 11.84APAGTTTTTTTTTRR54 pKa = 11.84TTRR57 pKa = 11.84TTGGRR62 pKa = 11.84HH63 pKa = 3.76GHH65 pKa = 6.51AAPVHH70 pKa = 4.63HH71 pKa = 7.34HH72 pKa = 6.34KK73 pKa = 10.63RR74 pKa = 11.84HH75 pKa = 4.81ATMGDD80 pKa = 3.45KK81 pKa = 10.91VSGMMMKK88 pKa = 10.48LRR90 pKa = 11.84GSLTRR95 pKa = 11.84RR96 pKa = 11.84PGLKK100 pKa = 10.1AAGTRR105 pKa = 11.84RR106 pKa = 11.84MHH108 pKa = 5.92GTDD111 pKa = 2.72GRR113 pKa = 11.84NARR116 pKa = 11.84RR117 pKa = 11.84VYY119 pKa = 10.89
Molecular weight: 13.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5528821 |
49 |
7213 |
430.0 |
47.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.763 ± 0.019 | 1.31 ± 0.009 |
5.583 ± 0.016 | 6.064 ± 0.021 |
3.695 ± 0.014 | 6.713 ± 0.021 |
2.483 ± 0.01 | 4.914 ± 0.016 |
5.1 ± 0.022 | 8.697 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.257 ± 0.008 | 3.778 ± 0.011 |
6.069 ± 0.025 | 4.15 ± 0.018 |
5.962 ± 0.019 | 8.044 ± 0.026 |
6.041 ± 0.014 | 6.043 ± 0.016 |
1.484 ± 0.009 | 2.85 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
