
Ophiophagus hannah (King cobra) (Naja hannah)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Lepidosauria;
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
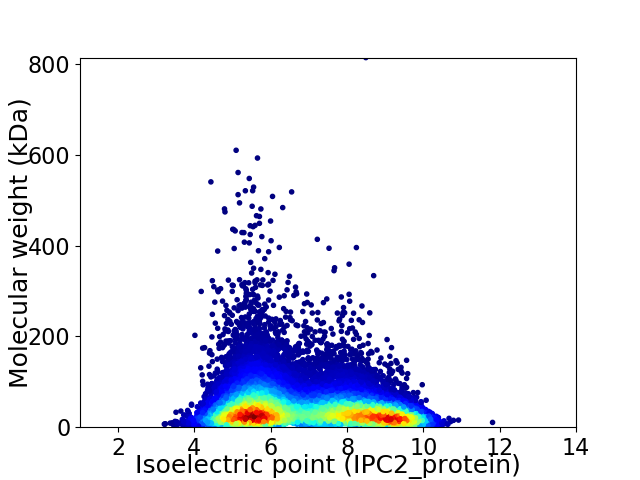
Virtual 2D-PAGE plot for 18387 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V8NVU6|V8NVU6_OPHHA Mastermind-like protein 2 (Fragment) OS=Ophiophagus hannah OX=8665 GN=MAML2 PE=4 SV=1
MM1 pKa = 7.49PQNCLSFQFNSDD13 pKa = 3.05SSEE16 pKa = 3.83EE17 pKa = 4.26GFGGNSSEE25 pKa = 4.27EE26 pKa = 4.16EE27 pKa = 4.09EE28 pKa = 4.39EE29 pKa = 4.42EE30 pKa = 4.63EE31 pKa = 4.48GGQSNEE37 pKa = 3.87EE38 pKa = 4.01NEE40 pKa = 4.28GGKK43 pKa = 10.49GSEE46 pKa = 4.33EE47 pKa = 4.05TTVAPTTPAAGDD59 pKa = 3.68TNTDD63 pKa = 3.38GNAEE67 pKa = 4.4PPTEE71 pKa = 4.2VPGVKK76 pKa = 10.24DD77 pKa = 3.62PLQKK81 pKa = 10.09PGKK84 pKa = 9.47EE85 pKa = 4.2GGDD88 pKa = 3.31KK89 pKa = 10.87KK90 pKa = 11.24VDD92 pKa = 3.41NAPKK96 pKa = 10.23KK97 pKa = 9.49GAKK100 pKa = 10.02EE101 pKa = 3.73EE102 pKa = 4.61DD103 pKa = 3.22SDD105 pKa = 4.36EE106 pKa = 4.54NEE108 pKa = 3.93EE109 pKa = 4.17EE110 pKa = 4.17EE111 pKa = 4.39NEE113 pKa = 4.09EE114 pKa = 3.99EE115 pKa = 4.23EE116 pKa = 4.45EE117 pKa = 4.33EE118 pKa = 4.37KK119 pKa = 10.95VDD121 pKa = 4.42EE122 pKa = 4.88NGSGANGTSTNSTLEE137 pKa = 3.82EE138 pKa = 4.04TGNGGEE144 pKa = 4.13EE145 pKa = 4.18EE146 pKa = 4.15EE147 pKa = 4.48EE148 pKa = 4.39EE149 pKa = 4.23EE150 pKa = 4.99NEE152 pKa = 3.89ATTISLTTLVTTVNPTEE169 pKa = 4.35STTGEE174 pKa = 3.69QWQDD178 pKa = 2.89GTSPADD184 pKa = 3.06QGQYY188 pKa = 9.07DD189 pKa = 4.07TEE191 pKa = 4.61DD192 pKa = 3.54PWVHH196 pKa = 7.29DD197 pKa = 3.55ATTTNGYY204 pKa = 8.85EE205 pKa = 4.12VQTDD209 pKa = 4.24YY210 pKa = 11.61YY211 pKa = 10.41GSEE214 pKa = 3.71NGYY217 pKa = 9.47PRR219 pKa = 11.84GDD221 pKa = 3.22NFRR224 pKa = 11.84TYY226 pKa = 10.58EE227 pKa = 4.32DD228 pKa = 3.88EE229 pKa = 3.79YY230 pKa = 11.4SYY232 pKa = 11.73YY233 pKa = 10.2KK234 pKa = 10.32GHH236 pKa = 6.87GYY238 pKa = 10.38DD239 pKa = 4.46VYY241 pKa = 11.4GQDD244 pKa = 3.42YY245 pKa = 10.51YY246 pKa = 12.03YY247 pKa = 11.17NQQ249 pKa = 3.36
MM1 pKa = 7.49PQNCLSFQFNSDD13 pKa = 3.05SSEE16 pKa = 3.83EE17 pKa = 4.26GFGGNSSEE25 pKa = 4.27EE26 pKa = 4.16EE27 pKa = 4.09EE28 pKa = 4.39EE29 pKa = 4.42EE30 pKa = 4.63EE31 pKa = 4.48GGQSNEE37 pKa = 3.87EE38 pKa = 4.01NEE40 pKa = 4.28GGKK43 pKa = 10.49GSEE46 pKa = 4.33EE47 pKa = 4.05TTVAPTTPAAGDD59 pKa = 3.68TNTDD63 pKa = 3.38GNAEE67 pKa = 4.4PPTEE71 pKa = 4.2VPGVKK76 pKa = 10.24DD77 pKa = 3.62PLQKK81 pKa = 10.09PGKK84 pKa = 9.47EE85 pKa = 4.2GGDD88 pKa = 3.31KK89 pKa = 10.87KK90 pKa = 11.24VDD92 pKa = 3.41NAPKK96 pKa = 10.23KK97 pKa = 9.49GAKK100 pKa = 10.02EE101 pKa = 3.73EE102 pKa = 4.61DD103 pKa = 3.22SDD105 pKa = 4.36EE106 pKa = 4.54NEE108 pKa = 3.93EE109 pKa = 4.17EE110 pKa = 4.17EE111 pKa = 4.39NEE113 pKa = 4.09EE114 pKa = 3.99EE115 pKa = 4.23EE116 pKa = 4.45EE117 pKa = 4.33EE118 pKa = 4.37KK119 pKa = 10.95VDD121 pKa = 4.42EE122 pKa = 4.88NGSGANGTSTNSTLEE137 pKa = 3.82EE138 pKa = 4.04TGNGGEE144 pKa = 4.13EE145 pKa = 4.18EE146 pKa = 4.15EE147 pKa = 4.48EE148 pKa = 4.39EE149 pKa = 4.23EE150 pKa = 4.99NEE152 pKa = 3.89ATTISLTTLVTTVNPTEE169 pKa = 4.35STTGEE174 pKa = 3.69QWQDD178 pKa = 2.89GTSPADD184 pKa = 3.06QGQYY188 pKa = 9.07DD189 pKa = 4.07TEE191 pKa = 4.61DD192 pKa = 3.54PWVHH196 pKa = 7.29DD197 pKa = 3.55ATTTNGYY204 pKa = 8.85EE205 pKa = 4.12VQTDD209 pKa = 4.24YY210 pKa = 11.61YY211 pKa = 10.41GSEE214 pKa = 3.71NGYY217 pKa = 9.47PRR219 pKa = 11.84GDD221 pKa = 3.22NFRR224 pKa = 11.84TYY226 pKa = 10.58EE227 pKa = 4.32DD228 pKa = 3.88EE229 pKa = 3.79YY230 pKa = 11.4SYY232 pKa = 11.73YY233 pKa = 10.2KK234 pKa = 10.32GHH236 pKa = 6.87GYY238 pKa = 10.38DD239 pKa = 4.46VYY241 pKa = 11.4GQDD244 pKa = 3.42YY245 pKa = 10.51YY246 pKa = 12.03YY247 pKa = 11.17NQQ249 pKa = 3.36
Molecular weight: 27.19 kDa
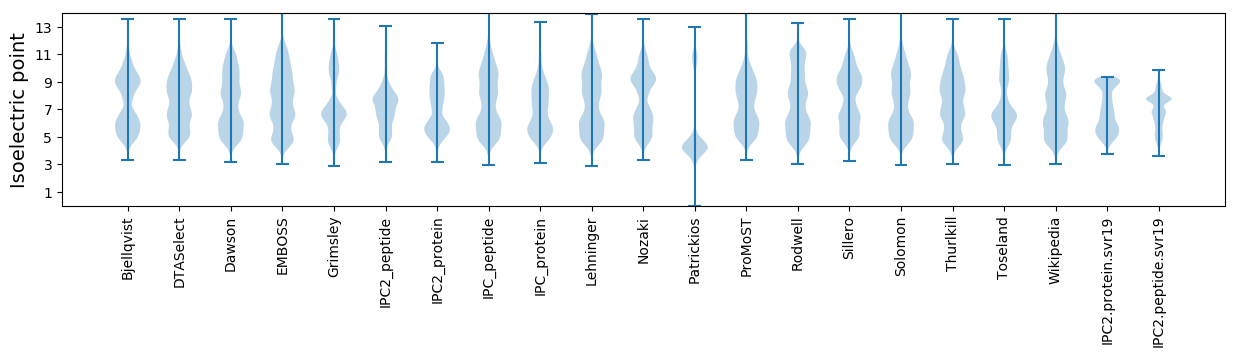
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V8P534|V8P534_OPHHA Polypyrimidine tract-binding protein 1 OS=Ophiophagus hannah OX=8665 GN=PTBP1 PE=4 SV=1
MM1 pKa = 7.48FSDD4 pKa = 3.81TVSYY8 pKa = 10.56KK9 pKa = 10.16VAIYY13 pKa = 8.43SHH15 pKa = 5.7WQARR19 pKa = 11.84TLGKK23 pKa = 10.43RR24 pKa = 11.84SDD26 pKa = 3.89FNVSGRR32 pKa = 11.84VVVVFEE38 pKa = 3.84GQYY41 pKa = 10.21RR42 pKa = 11.84DD43 pKa = 3.06SFLEE47 pKa = 3.81LRR49 pKa = 11.84ARR51 pKa = 11.84TANSRR56 pKa = 11.84YY57 pKa = 9.52FLGAEE62 pKa = 4.28STRR65 pKa = 11.84QFPSLSDD72 pKa = 3.11STNRR76 pKa = 11.84VISEE80 pKa = 4.02SAGYY84 pKa = 8.1EE85 pKa = 3.8LKK87 pKa = 11.06VRR89 pKa = 11.84MKK91 pKa = 10.57VAAKK95 pKa = 10.12PIGEE99 pKa = 4.42CWCVCAQSTNWGCRR113 pKa = 11.84FRR115 pKa = 11.84LFWPGYY121 pKa = 8.4GASPPPRR128 pKa = 11.84HH129 pKa = 6.36GSPCPCSSPLSEE141 pKa = 4.06RR142 pKa = 11.84NRR144 pKa = 11.84RR145 pKa = 11.84LLLSSNSAAAILQRR159 pKa = 11.84QRR161 pKa = 11.84SKK163 pKa = 10.91GRR165 pKa = 11.84EE166 pKa = 3.61AGRR169 pKa = 11.84EE170 pKa = 3.88KK171 pKa = 10.73RR172 pKa = 11.84GGQLKK177 pKa = 9.45TYY179 pKa = 10.03SRR181 pKa = 11.84PKK183 pKa = 9.21PSYY186 pKa = 9.65ARR188 pKa = 11.84RR189 pKa = 11.84WQQSGDD195 pKa = 3.98PLDD198 pKa = 3.82QAAGSAPVRR207 pKa = 11.84PLLRR211 pKa = 11.84NPPPVTPAAGPTLKK225 pKa = 10.03GTGLGRR231 pKa = 11.84VLYY234 pKa = 10.64SGGLPFPLSKK244 pKa = 10.85ASAGRR249 pKa = 11.84GRR251 pKa = 11.84FLRR254 pKa = 11.84RR255 pKa = 11.84CLATSGFGQHH265 pKa = 6.42AVSEE269 pKa = 4.66CVCVSVCVCACVCVGHH285 pKa = 5.7TQSTVTRR292 pKa = 11.84AHH294 pKa = 6.85LAWSMPGNGKK304 pKa = 10.03RR305 pKa = 11.84GSRR308 pKa = 11.84QAA310 pKa = 3.44
MM1 pKa = 7.48FSDD4 pKa = 3.81TVSYY8 pKa = 10.56KK9 pKa = 10.16VAIYY13 pKa = 8.43SHH15 pKa = 5.7WQARR19 pKa = 11.84TLGKK23 pKa = 10.43RR24 pKa = 11.84SDD26 pKa = 3.89FNVSGRR32 pKa = 11.84VVVVFEE38 pKa = 3.84GQYY41 pKa = 10.21RR42 pKa = 11.84DD43 pKa = 3.06SFLEE47 pKa = 3.81LRR49 pKa = 11.84ARR51 pKa = 11.84TANSRR56 pKa = 11.84YY57 pKa = 9.52FLGAEE62 pKa = 4.28STRR65 pKa = 11.84QFPSLSDD72 pKa = 3.11STNRR76 pKa = 11.84VISEE80 pKa = 4.02SAGYY84 pKa = 8.1EE85 pKa = 3.8LKK87 pKa = 11.06VRR89 pKa = 11.84MKK91 pKa = 10.57VAAKK95 pKa = 10.12PIGEE99 pKa = 4.42CWCVCAQSTNWGCRR113 pKa = 11.84FRR115 pKa = 11.84LFWPGYY121 pKa = 8.4GASPPPRR128 pKa = 11.84HH129 pKa = 6.36GSPCPCSSPLSEE141 pKa = 4.06RR142 pKa = 11.84NRR144 pKa = 11.84RR145 pKa = 11.84LLLSSNSAAAILQRR159 pKa = 11.84QRR161 pKa = 11.84SKK163 pKa = 10.91GRR165 pKa = 11.84EE166 pKa = 3.61AGRR169 pKa = 11.84EE170 pKa = 3.88KK171 pKa = 10.73RR172 pKa = 11.84GGQLKK177 pKa = 9.45TYY179 pKa = 10.03SRR181 pKa = 11.84PKK183 pKa = 9.21PSYY186 pKa = 9.65ARR188 pKa = 11.84RR189 pKa = 11.84WQQSGDD195 pKa = 3.98PLDD198 pKa = 3.82QAAGSAPVRR207 pKa = 11.84PLLRR211 pKa = 11.84NPPPVTPAAGPTLKK225 pKa = 10.03GTGLGRR231 pKa = 11.84VLYY234 pKa = 10.64SGGLPFPLSKK244 pKa = 10.85ASAGRR249 pKa = 11.84GRR251 pKa = 11.84FLRR254 pKa = 11.84RR255 pKa = 11.84CLATSGFGQHH265 pKa = 6.42AVSEE269 pKa = 4.66CVCVSVCVCACVCVGHH285 pKa = 5.7TQSTVTRR292 pKa = 11.84AHH294 pKa = 6.85LAWSMPGNGKK304 pKa = 10.03RR305 pKa = 11.84GSRR308 pKa = 11.84QAA310 pKa = 3.44
Molecular weight: 33.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7683767 |
10 |
7049 |
417.9 |
46.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.144 ± 0.016 | 2.215 ± 0.015 |
4.726 ± 0.015 | 7.526 ± 0.029 |
3.813 ± 0.015 | 6.321 ± 0.029 |
2.557 ± 0.009 | 4.972 ± 0.017 |
6.641 ± 0.026 | 9.65 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.276 ± 0.008 | 4.129 ± 0.016 |
5.538 ± 0.024 | 4.705 ± 0.018 |
5.615 ± 0.022 | 8.257 ± 0.023 |
5.216 ± 0.014 | 5.709 ± 0.015 |
1.251 ± 0.007 | 2.724 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
