
Bosea sp. Root381
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Boseaceae; Bosea; unclassified Bosea
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
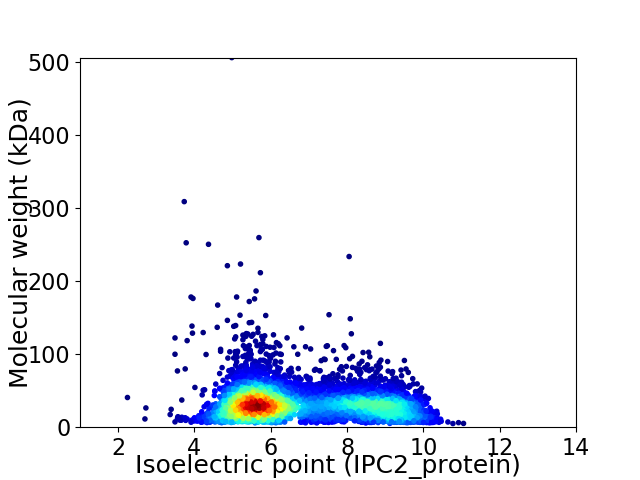
Virtual 2D-PAGE plot for 5096 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q9IME3|A0A0Q9IME3_9BRAD Peptidase M16 OS=Bosea sp. Root381 OX=1736524 GN=ASE63_02205 PE=3 SV=1
MM1 pKa = 7.47TNLTIYY7 pKa = 10.23AHH9 pKa = 7.33RR10 pKa = 11.84GTNPYY15 pKa = 9.12PDD17 pKa = 4.25HH18 pKa = 7.4SRR20 pKa = 11.84DD21 pKa = 3.54AYY23 pKa = 10.05VWAVNYY29 pKa = 10.01GADD32 pKa = 4.56FIEE35 pKa = 5.21PDD37 pKa = 4.04LFLTKK42 pKa = 10.54DD43 pKa = 3.49GVLVASHH50 pKa = 7.49DD51 pKa = 3.64NHH53 pKa = 6.56NYY55 pKa = 10.82ANLTYY60 pKa = 10.62AEE62 pKa = 4.38AKK64 pKa = 10.32AIEE67 pKa = 4.33PALMTFGEE75 pKa = 4.56VIEE78 pKa = 4.7IAKK81 pKa = 10.0QMSIEE86 pKa = 4.14TGRR89 pKa = 11.84QIGVIPEE96 pKa = 4.24TKK98 pKa = 9.4SANYY102 pKa = 7.54ATSEE106 pKa = 3.93AVIRR110 pKa = 11.84DD111 pKa = 4.54LIAHH115 pKa = 7.39DD116 pKa = 3.95FTDD119 pKa = 4.04PNLVVIQSFQSSNLKK134 pKa = 8.86MLHH137 pKa = 5.04EE138 pKa = 5.04TIMPQYY144 pKa = 10.95GVDD147 pKa = 3.83LPLAFLGYY155 pKa = 10.6NMSAATIADD164 pKa = 3.73TATYY168 pKa = 11.04ADD170 pKa = 4.57IIAPNQAALTAAGIEE185 pKa = 4.1AAHH188 pKa = 6.27AAGLKK193 pKa = 9.28VVTWTVLGTEE203 pKa = 4.12AQIQRR208 pKa = 11.84LVDD211 pKa = 4.08LGVDD215 pKa = 3.85GVFVDD220 pKa = 4.02ATNTARR226 pKa = 11.84EE227 pKa = 4.29ALSKK231 pKa = 10.88INGVTVGYY239 pKa = 8.46GTEE242 pKa = 4.34GDD244 pKa = 4.28DD245 pKa = 5.06EE246 pKa = 4.46IAGTDD251 pKa = 3.96GDD253 pKa = 4.09DD254 pKa = 5.34LIYY257 pKa = 11.13GMAGDD262 pKa = 5.05DD263 pKa = 4.49EE264 pKa = 5.21ISAGDD269 pKa = 3.91GNDD272 pKa = 3.2VVYY275 pKa = 11.03GDD277 pKa = 4.76AGDD280 pKa = 5.1DD281 pKa = 3.86IIEE284 pKa = 4.54GGAGNDD290 pKa = 3.55VLVGGAGDD298 pKa = 4.29DD299 pKa = 3.88EE300 pKa = 5.22LFGGAGDD307 pKa = 4.45DD308 pKa = 3.75VLKK311 pKa = 11.2GGVGDD316 pKa = 4.78DD317 pKa = 4.53LLDD320 pKa = 4.34GGDD323 pKa = 4.15GVDD326 pKa = 3.22TADD329 pKa = 4.67YY330 pKa = 11.13SDD332 pKa = 4.09DD333 pKa = 3.88TAGVTVDD340 pKa = 4.58LSAGTASGDD349 pKa = 3.53EE350 pKa = 4.41AGDD353 pKa = 4.2DD354 pKa = 3.87EE355 pKa = 6.22LISS358 pKa = 3.69
MM1 pKa = 7.47TNLTIYY7 pKa = 10.23AHH9 pKa = 7.33RR10 pKa = 11.84GTNPYY15 pKa = 9.12PDD17 pKa = 4.25HH18 pKa = 7.4SRR20 pKa = 11.84DD21 pKa = 3.54AYY23 pKa = 10.05VWAVNYY29 pKa = 10.01GADD32 pKa = 4.56FIEE35 pKa = 5.21PDD37 pKa = 4.04LFLTKK42 pKa = 10.54DD43 pKa = 3.49GVLVASHH50 pKa = 7.49DD51 pKa = 3.64NHH53 pKa = 6.56NYY55 pKa = 10.82ANLTYY60 pKa = 10.62AEE62 pKa = 4.38AKK64 pKa = 10.32AIEE67 pKa = 4.33PALMTFGEE75 pKa = 4.56VIEE78 pKa = 4.7IAKK81 pKa = 10.0QMSIEE86 pKa = 4.14TGRR89 pKa = 11.84QIGVIPEE96 pKa = 4.24TKK98 pKa = 9.4SANYY102 pKa = 7.54ATSEE106 pKa = 3.93AVIRR110 pKa = 11.84DD111 pKa = 4.54LIAHH115 pKa = 7.39DD116 pKa = 3.95FTDD119 pKa = 4.04PNLVVIQSFQSSNLKK134 pKa = 8.86MLHH137 pKa = 5.04EE138 pKa = 5.04TIMPQYY144 pKa = 10.95GVDD147 pKa = 3.83LPLAFLGYY155 pKa = 10.6NMSAATIADD164 pKa = 3.73TATYY168 pKa = 11.04ADD170 pKa = 4.57IIAPNQAALTAAGIEE185 pKa = 4.1AAHH188 pKa = 6.27AAGLKK193 pKa = 9.28VVTWTVLGTEE203 pKa = 4.12AQIQRR208 pKa = 11.84LVDD211 pKa = 4.08LGVDD215 pKa = 3.85GVFVDD220 pKa = 4.02ATNTARR226 pKa = 11.84EE227 pKa = 4.29ALSKK231 pKa = 10.88INGVTVGYY239 pKa = 8.46GTEE242 pKa = 4.34GDD244 pKa = 4.28DD245 pKa = 5.06EE246 pKa = 4.46IAGTDD251 pKa = 3.96GDD253 pKa = 4.09DD254 pKa = 5.34LIYY257 pKa = 11.13GMAGDD262 pKa = 5.05DD263 pKa = 4.49EE264 pKa = 5.21ISAGDD269 pKa = 3.91GNDD272 pKa = 3.2VVYY275 pKa = 11.03GDD277 pKa = 4.76AGDD280 pKa = 5.1DD281 pKa = 3.86IIEE284 pKa = 4.54GGAGNDD290 pKa = 3.55VLVGGAGDD298 pKa = 4.29DD299 pKa = 3.88EE300 pKa = 5.22LFGGAGDD307 pKa = 4.45DD308 pKa = 3.75VLKK311 pKa = 11.2GGVGDD316 pKa = 4.78DD317 pKa = 4.53LLDD320 pKa = 4.34GGDD323 pKa = 4.15GVDD326 pKa = 3.22TADD329 pKa = 4.67YY330 pKa = 11.13SDD332 pKa = 4.09DD333 pKa = 3.88TAGVTVDD340 pKa = 4.58LSAGTASGDD349 pKa = 3.53EE350 pKa = 4.41AGDD353 pKa = 4.2DD354 pKa = 3.87EE355 pKa = 6.22LISS358 pKa = 3.69
Molecular weight: 37.18 kDa
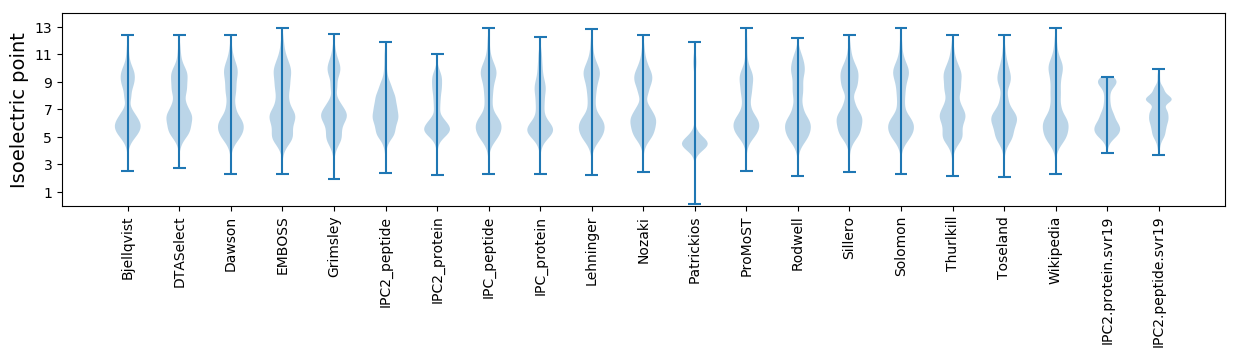
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q9I4U9|A0A0Q9I4U9_9BRAD Chaperone protein DnaK OS=Bosea sp. Root381 OX=1736524 GN=dnaK PE=2 SV=1
MM1 pKa = 7.69LSPIRR6 pKa = 11.84AAGCKK11 pKa = 9.13RR12 pKa = 11.84WPCGAEE18 pKa = 3.32PAARR22 pKa = 11.84HH23 pKa = 4.95TKK25 pKa = 9.1RR26 pKa = 11.84TFSDD30 pKa = 2.96NAADD34 pKa = 3.5KK35 pKa = 10.92RR36 pKa = 11.84VGQEE40 pKa = 3.57ALRR43 pKa = 11.84AMIQIKK49 pKa = 10.04GPRR52 pKa = 11.84DD53 pKa = 3.32FAAGCTFILIALFYY67 pKa = 10.67LWFGRR72 pKa = 11.84NLSIGSSSFMEE83 pKa = 4.14AGYY86 pKa = 9.8FPRR89 pKa = 11.84LAAVALALFGAIIAVRR105 pKa = 11.84AVAIDD110 pKa = 4.07GPGLEE115 pKa = 4.12PWAWRR120 pKa = 11.84PLLILTGCVVLFGLIITRR138 pKa = 11.84AGLVLTTVLVVVAMSFAGGRR158 pKa = 11.84LSWTRR163 pKa = 11.84LAGFVAALVVFMVGLFHH180 pKa = 7.51FGLGLAIPVWPQQ192 pKa = 2.47
MM1 pKa = 7.69LSPIRR6 pKa = 11.84AAGCKK11 pKa = 9.13RR12 pKa = 11.84WPCGAEE18 pKa = 3.32PAARR22 pKa = 11.84HH23 pKa = 4.95TKK25 pKa = 9.1RR26 pKa = 11.84TFSDD30 pKa = 2.96NAADD34 pKa = 3.5KK35 pKa = 10.92RR36 pKa = 11.84VGQEE40 pKa = 3.57ALRR43 pKa = 11.84AMIQIKK49 pKa = 10.04GPRR52 pKa = 11.84DD53 pKa = 3.32FAAGCTFILIALFYY67 pKa = 10.67LWFGRR72 pKa = 11.84NLSIGSSSFMEE83 pKa = 4.14AGYY86 pKa = 9.8FPRR89 pKa = 11.84LAAVALALFGAIIAVRR105 pKa = 11.84AVAIDD110 pKa = 4.07GPGLEE115 pKa = 4.12PWAWRR120 pKa = 11.84PLLILTGCVVLFGLIITRR138 pKa = 11.84AGLVLTTVLVVVAMSFAGGRR158 pKa = 11.84LSWTRR163 pKa = 11.84LAGFVAALVVFMVGLFHH180 pKa = 7.51FGLGLAIPVWPQQ192 pKa = 2.47
Molecular weight: 20.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1611244 |
41 |
5166 |
316.2 |
34.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.498 ± 0.045 | 0.781 ± 0.011 |
5.361 ± 0.03 | 5.553 ± 0.033 |
3.672 ± 0.023 | 9.064 ± 0.06 |
1.896 ± 0.016 | 5.096 ± 0.024 |
3.069 ± 0.03 | 10.431 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.02 | 2.354 ± 0.028 |
5.352 ± 0.034 | 3.025 ± 0.016 |
7.232 ± 0.04 | 5.294 ± 0.033 |
5.193 ± 0.037 | 7.448 ± 0.031 |
1.295 ± 0.017 | 2.041 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
