
Wuhan insect virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
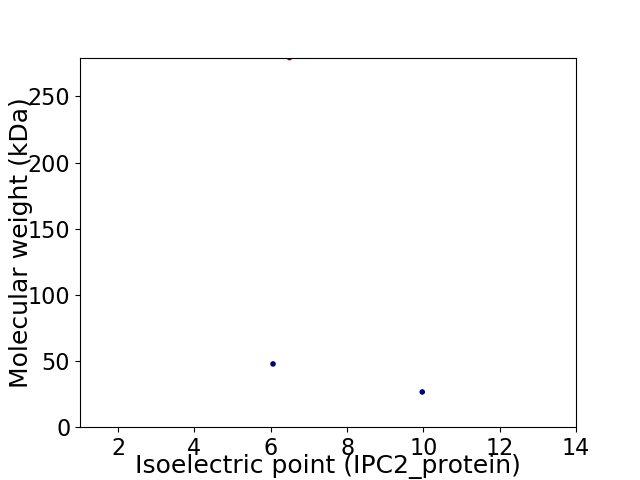
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KKE4|A0A1L3KKE4_9VIRU Helicase OS=Wuhan insect virus 9 OX=1923740 PE=4 SV=1
MM1 pKa = 7.91CYY3 pKa = 10.29FILILYY9 pKa = 7.35LTHH12 pKa = 7.05IAHH15 pKa = 7.28GAPVNTKK22 pKa = 7.17TTTLRR27 pKa = 11.84NLNITFPGIFQHH39 pKa = 6.55NFLDD43 pKa = 4.62TIDD46 pKa = 5.25FYY48 pKa = 11.18PSNKK52 pKa = 7.49YY53 pKa = 6.98TQFIAPKK60 pKa = 8.55FQSKK64 pKa = 10.08CMYY67 pKa = 10.64ASVEE71 pKa = 4.04NFLTEE76 pKa = 4.05SNYY79 pKa = 10.19VMPQEE84 pKa = 4.89CGLVATMTIHH94 pKa = 8.0DD95 pKa = 4.17SMFTKK100 pKa = 10.53SIVDD104 pKa = 3.45VGTFACTQYY113 pKa = 10.78HH114 pKa = 6.16RR115 pKa = 11.84NKK117 pKa = 10.03CNPPMHH123 pKa = 7.39LYY125 pKa = 8.62TLNYY129 pKa = 9.61TEE131 pKa = 5.13DD132 pKa = 3.7AFVNDD137 pKa = 4.12IITYY141 pKa = 8.16YY142 pKa = 10.64SKK144 pKa = 9.87EE145 pKa = 4.44TITFRR150 pKa = 11.84YY151 pKa = 7.84TLDD154 pKa = 3.29TTLINYY160 pKa = 9.63IIVLKK165 pKa = 10.82DD166 pKa = 3.17EE167 pKa = 4.38YY168 pKa = 11.03TGMFYY173 pKa = 10.17WITSYY178 pKa = 10.62CLKK181 pKa = 10.73YY182 pKa = 8.34MTVTSDD188 pKa = 2.87NKK190 pKa = 10.0FVHH193 pKa = 6.82LGPGIHH199 pKa = 7.01HH200 pKa = 6.93YY201 pKa = 10.6DD202 pKa = 3.19AATKK206 pKa = 8.94QFCITDD212 pKa = 3.63YY213 pKa = 11.73SNMCTPVGPFKK224 pKa = 10.78TKK226 pKa = 9.78IDD228 pKa = 4.57FILSLPLEE236 pKa = 4.25ITISSDD242 pKa = 3.2RR243 pKa = 11.84PYY245 pKa = 10.59HH246 pKa = 6.79KK247 pKa = 10.59YY248 pKa = 10.34FEE250 pKa = 4.22KK251 pKa = 9.63HH252 pKa = 5.74TYY254 pKa = 10.73DD255 pKa = 3.42DD256 pKa = 4.38TVKK259 pKa = 9.48HH260 pKa = 6.22TIPYY264 pKa = 9.87DD265 pKa = 3.59DD266 pKa = 3.83NNRR269 pKa = 11.84FEE271 pKa = 4.54FDD273 pKa = 3.55KK274 pKa = 11.04NDD276 pKa = 3.26HH277 pKa = 5.8FVYY280 pKa = 10.52VAGNIRR286 pKa = 11.84SMQPNYY292 pKa = 10.5QCVKK296 pKa = 7.79YY297 pKa = 9.74HH298 pKa = 5.35YY299 pKa = 10.38VPMSSIFHH307 pKa = 6.75IDD309 pKa = 3.53IIIDD313 pKa = 3.95SIEE316 pKa = 4.22KK317 pKa = 10.66KK318 pKa = 6.87MTQYY322 pKa = 11.06FHH324 pKa = 7.45LLTNFINDD332 pKa = 3.22NVHH335 pKa = 6.73IIVEE339 pKa = 4.43HH340 pKa = 6.57IEE342 pKa = 4.22NIVLVTCSYY351 pKa = 10.02VVKK354 pKa = 10.56KK355 pKa = 10.17ILYY358 pKa = 9.71LITRR362 pKa = 11.84ILAYY366 pKa = 10.13SYY368 pKa = 10.96IMEE371 pKa = 4.65AFAITMYY378 pKa = 10.87MMITNYY384 pKa = 10.51NYY386 pKa = 7.72TTIACVVATFCILKK400 pKa = 9.84KK401 pKa = 9.9WLFMWW406 pKa = 4.73
MM1 pKa = 7.91CYY3 pKa = 10.29FILILYY9 pKa = 7.35LTHH12 pKa = 7.05IAHH15 pKa = 7.28GAPVNTKK22 pKa = 7.17TTTLRR27 pKa = 11.84NLNITFPGIFQHH39 pKa = 6.55NFLDD43 pKa = 4.62TIDD46 pKa = 5.25FYY48 pKa = 11.18PSNKK52 pKa = 7.49YY53 pKa = 6.98TQFIAPKK60 pKa = 8.55FQSKK64 pKa = 10.08CMYY67 pKa = 10.64ASVEE71 pKa = 4.04NFLTEE76 pKa = 4.05SNYY79 pKa = 10.19VMPQEE84 pKa = 4.89CGLVATMTIHH94 pKa = 8.0DD95 pKa = 4.17SMFTKK100 pKa = 10.53SIVDD104 pKa = 3.45VGTFACTQYY113 pKa = 10.78HH114 pKa = 6.16RR115 pKa = 11.84NKK117 pKa = 10.03CNPPMHH123 pKa = 7.39LYY125 pKa = 8.62TLNYY129 pKa = 9.61TEE131 pKa = 5.13DD132 pKa = 3.7AFVNDD137 pKa = 4.12IITYY141 pKa = 8.16YY142 pKa = 10.64SKK144 pKa = 9.87EE145 pKa = 4.44TITFRR150 pKa = 11.84YY151 pKa = 7.84TLDD154 pKa = 3.29TTLINYY160 pKa = 9.63IIVLKK165 pKa = 10.82DD166 pKa = 3.17EE167 pKa = 4.38YY168 pKa = 11.03TGMFYY173 pKa = 10.17WITSYY178 pKa = 10.62CLKK181 pKa = 10.73YY182 pKa = 8.34MTVTSDD188 pKa = 2.87NKK190 pKa = 10.0FVHH193 pKa = 6.82LGPGIHH199 pKa = 7.01HH200 pKa = 6.93YY201 pKa = 10.6DD202 pKa = 3.19AATKK206 pKa = 8.94QFCITDD212 pKa = 3.63YY213 pKa = 11.73SNMCTPVGPFKK224 pKa = 10.78TKK226 pKa = 9.78IDD228 pKa = 4.57FILSLPLEE236 pKa = 4.25ITISSDD242 pKa = 3.2RR243 pKa = 11.84PYY245 pKa = 10.59HH246 pKa = 6.79KK247 pKa = 10.59YY248 pKa = 10.34FEE250 pKa = 4.22KK251 pKa = 9.63HH252 pKa = 5.74TYY254 pKa = 10.73DD255 pKa = 3.42DD256 pKa = 4.38TVKK259 pKa = 9.48HH260 pKa = 6.22TIPYY264 pKa = 9.87DD265 pKa = 3.59DD266 pKa = 3.83NNRR269 pKa = 11.84FEE271 pKa = 4.54FDD273 pKa = 3.55KK274 pKa = 11.04NDD276 pKa = 3.26HH277 pKa = 5.8FVYY280 pKa = 10.52VAGNIRR286 pKa = 11.84SMQPNYY292 pKa = 10.5QCVKK296 pKa = 7.79YY297 pKa = 9.74HH298 pKa = 5.35YY299 pKa = 10.38VPMSSIFHH307 pKa = 6.75IDD309 pKa = 3.53IIIDD313 pKa = 3.95SIEE316 pKa = 4.22KK317 pKa = 10.66KK318 pKa = 6.87MTQYY322 pKa = 11.06FHH324 pKa = 7.45LLTNFINDD332 pKa = 3.22NVHH335 pKa = 6.73IIVEE339 pKa = 4.43HH340 pKa = 6.57IEE342 pKa = 4.22NIVLVTCSYY351 pKa = 10.02VVKK354 pKa = 10.56KK355 pKa = 10.17ILYY358 pKa = 9.71LITRR362 pKa = 11.84ILAYY366 pKa = 10.13SYY368 pKa = 10.96IMEE371 pKa = 4.65AFAITMYY378 pKa = 10.87MMITNYY384 pKa = 10.51NYY386 pKa = 7.72TTIACVVATFCILKK400 pKa = 9.84KK401 pKa = 9.9WLFMWW406 pKa = 4.73
Molecular weight: 47.81 kDa
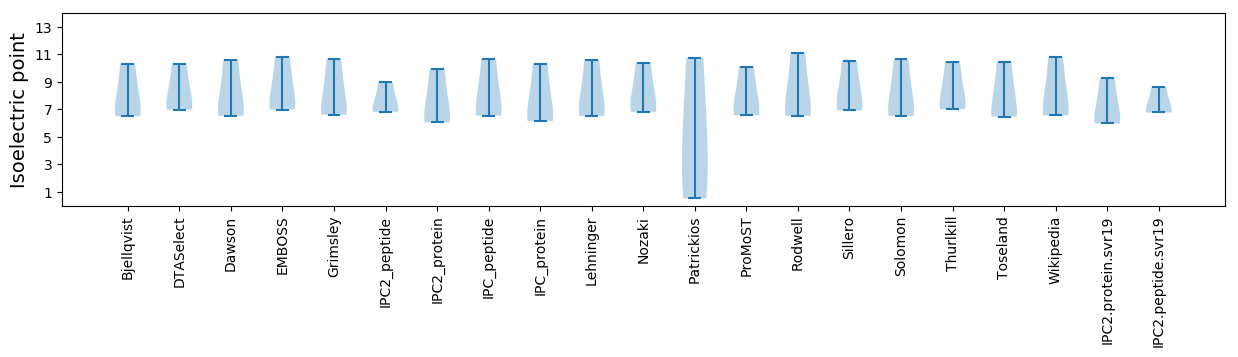
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKB7|A0A1L3KKB7_9VIRU Uncharacterized protein OS=Wuhan insect virus 9 OX=1923740 PE=4 SV=1
MM1 pKa = 7.91PKK3 pKa = 9.34YY4 pKa = 10.26QKK6 pKa = 9.84TITFVNKK13 pKa = 9.97KK14 pKa = 9.13YY15 pKa = 10.65KK16 pKa = 10.13NRR18 pKa = 11.84NALPMKK24 pKa = 9.47PRR26 pKa = 11.84RR27 pKa = 11.84SRR29 pKa = 11.84SRR31 pKa = 11.84TPSVRR36 pKa = 11.84RR37 pKa = 11.84SRR39 pKa = 11.84SRR41 pKa = 11.84SRR43 pKa = 11.84SRR45 pKa = 11.84TPKK48 pKa = 9.42RR49 pKa = 11.84TTIKK53 pKa = 10.59YY54 pKa = 9.34GVVDD58 pKa = 3.78GFINRR63 pKa = 11.84IMKK66 pKa = 9.93KK67 pKa = 10.06FSSILVDD74 pKa = 4.06PYY76 pKa = 11.2TYY78 pKa = 10.15IGLALMSLLMYY89 pKa = 10.07IHH91 pKa = 6.63YY92 pKa = 9.67FHH94 pKa = 7.68IDD96 pKa = 3.36DD97 pKa = 4.18SAVNSIVKK105 pKa = 8.44TLKK108 pKa = 10.96KK109 pKa = 10.29NDD111 pKa = 3.49STKK114 pKa = 11.11AFADD118 pKa = 3.44WVEE121 pKa = 4.03GNINKK126 pKa = 9.17IFAIVMLLYY135 pKa = 10.5HH136 pKa = 6.7SLQIPIKK143 pKa = 9.33YY144 pKa = 9.87RR145 pKa = 11.84YY146 pKa = 8.07FTILLSMITVFLMPSLSVVTYY167 pKa = 9.13SATIFGVVMYY177 pKa = 10.44HH178 pKa = 6.57KK179 pKa = 9.92MKK181 pKa = 10.63KK182 pKa = 8.88RR183 pKa = 11.84TDD185 pKa = 2.99KK186 pKa = 11.19FIILLFYY193 pKa = 10.19IIIMSWFYY201 pKa = 10.37YY202 pKa = 10.64DD203 pKa = 5.52EE204 pKa = 4.81IIKK207 pKa = 9.88YY208 pKa = 9.48HH209 pKa = 6.28SKK211 pKa = 5.56TTRR214 pKa = 11.84VKK216 pKa = 10.83RR217 pKa = 11.84EE218 pKa = 3.85TEE220 pKa = 3.96EE221 pKa = 3.92QQVV224 pKa = 3.07
MM1 pKa = 7.91PKK3 pKa = 9.34YY4 pKa = 10.26QKK6 pKa = 9.84TITFVNKK13 pKa = 9.97KK14 pKa = 9.13YY15 pKa = 10.65KK16 pKa = 10.13NRR18 pKa = 11.84NALPMKK24 pKa = 9.47PRR26 pKa = 11.84RR27 pKa = 11.84SRR29 pKa = 11.84SRR31 pKa = 11.84TPSVRR36 pKa = 11.84RR37 pKa = 11.84SRR39 pKa = 11.84SRR41 pKa = 11.84SRR43 pKa = 11.84SRR45 pKa = 11.84TPKK48 pKa = 9.42RR49 pKa = 11.84TTIKK53 pKa = 10.59YY54 pKa = 9.34GVVDD58 pKa = 3.78GFINRR63 pKa = 11.84IMKK66 pKa = 9.93KK67 pKa = 10.06FSSILVDD74 pKa = 4.06PYY76 pKa = 11.2TYY78 pKa = 10.15IGLALMSLLMYY89 pKa = 10.07IHH91 pKa = 6.63YY92 pKa = 9.67FHH94 pKa = 7.68IDD96 pKa = 3.36DD97 pKa = 4.18SAVNSIVKK105 pKa = 8.44TLKK108 pKa = 10.96KK109 pKa = 10.29NDD111 pKa = 3.49STKK114 pKa = 11.11AFADD118 pKa = 3.44WVEE121 pKa = 4.03GNINKK126 pKa = 9.17IFAIVMLLYY135 pKa = 10.5HH136 pKa = 6.7SLQIPIKK143 pKa = 9.33YY144 pKa = 9.87RR145 pKa = 11.84YY146 pKa = 8.07FTILLSMITVFLMPSLSVVTYY167 pKa = 9.13SATIFGVVMYY177 pKa = 10.44HH178 pKa = 6.57KK179 pKa = 9.92MKK181 pKa = 10.63KK182 pKa = 8.88RR183 pKa = 11.84TDD185 pKa = 2.99KK186 pKa = 11.19FIILLFYY193 pKa = 10.19IIIMSWFYY201 pKa = 10.37YY202 pKa = 10.64DD203 pKa = 5.52EE204 pKa = 4.81IIKK207 pKa = 9.88YY208 pKa = 9.48HH209 pKa = 6.28SKK211 pKa = 5.56TTRR214 pKa = 11.84VKK216 pKa = 10.83RR217 pKa = 11.84EE218 pKa = 3.85TEE220 pKa = 3.96EE221 pKa = 3.92QQVV224 pKa = 3.07
Molecular weight: 26.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3048 |
224 |
2418 |
1016.0 |
117.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.199 ± 0.364 | 1.739 ± 0.442 |
7.283 ± 1.287 | 4.199 ± 0.684 |
4.79 ± 0.674 | 3.051 ± 0.399 |
2.953 ± 0.437 | 8.071 ± 1.239 |
8.399 ± 0.742 | 7.415 ± 0.361 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.707 ± 0.387 | 5.381 ± 0.407 |
3.051 ± 0.287 | 2.329 ± 0.152 |
3.675 ± 0.899 | 7.907 ± 0.969 |
7.283 ± 1.108 | 7.087 ± 0.376 |
0.623 ± 0.089 | 6.857 ± 0.606 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
