
Flavobacterium pallidum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
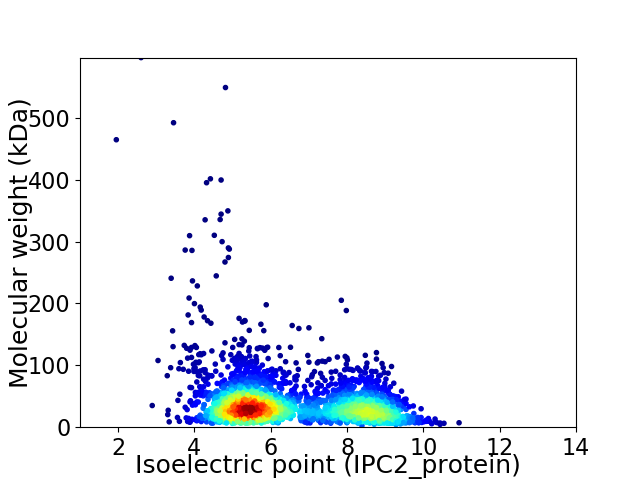
Virtual 2D-PAGE plot for 2931 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S1SHV9|A0A2S1SHV9_9FLAO Cyanophycin synthase OS=Flavobacterium pallidum OX=2172098 GN=cphA PE=3 SV=1
MM1 pKa = 7.59KK2 pKa = 10.51KK3 pKa = 10.23NLLFLIALICFSLGNAQSISIVGEE27 pKa = 4.0AVGGWEE33 pKa = 4.95DD34 pKa = 4.23NDD36 pKa = 4.09PNTPDD41 pKa = 3.19THH43 pKa = 8.76IMSTVDD49 pKa = 3.31NINYY53 pKa = 7.51TFSNLAVATADD64 pKa = 3.89PNGGGAKK71 pKa = 9.79FRR73 pKa = 11.84QDD75 pKa = 3.37GAWAINWGNAAFPSGTGTQNGANILTVAGTYY106 pKa = 10.55DD107 pKa = 3.26VTFNRR112 pKa = 11.84LTGAYY117 pKa = 9.96SFTNVVTYY125 pKa = 8.94PSIGMIGTAVNANGFNGPDD144 pKa = 3.05VDD146 pKa = 4.19MVTFDD151 pKa = 4.49GITYY155 pKa = 8.38TITNYY160 pKa = 10.43NFLSGEE166 pKa = 4.1MKK168 pKa = 10.44FRR170 pKa = 11.84LDD172 pKa = 4.85DD173 pKa = 3.45SWDD176 pKa = 3.72NSWGSTAFPSGTAVLNVPDD195 pKa = 5.49NIPVPAGTYY204 pKa = 8.9SVSFNRR210 pKa = 11.84ITGDD214 pKa = 3.32YY215 pKa = 10.37SFTFPSIGIIGTAVSAGGFNDD236 pKa = 3.97PDD238 pKa = 4.26TDD240 pKa = 4.4LMTADD245 pKa = 3.34GVHH248 pKa = 5.25YY249 pKa = 10.55VLYY252 pKa = 10.74NHH254 pKa = 6.21VFTDD258 pKa = 4.12GLAKK262 pKa = 10.41FRR264 pKa = 11.84QGDD267 pKa = 3.3NWDD270 pKa = 4.16VNWGNADD277 pKa = 3.25GDD279 pKa = 4.19FPSGIGTQGGTDD291 pKa = 3.18IPVTAGTYY299 pKa = 7.21TVNFNRR305 pKa = 11.84EE306 pKa = 3.37TGAYY310 pKa = 9.95SFDD313 pKa = 3.78APVVFANIGVIGTAVSAGGFDD334 pKa = 5.84DD335 pKa = 5.91PDD337 pKa = 3.58TDD339 pKa = 3.39MTTTDD344 pKa = 4.33GITYY348 pKa = 8.93TLEE351 pKa = 3.95NYY353 pKa = 9.5VFANGQLKK361 pKa = 9.77FRR363 pKa = 11.84QDD365 pKa = 4.12DD366 pKa = 3.56VWDD369 pKa = 4.13NNWGGVGFPGDD380 pKa = 3.37TANFNGPNIPVTAGTYY396 pKa = 9.2TLTFNINTLAYY407 pKa = 9.84SFTGTPLYY415 pKa = 8.21PTVGILGTAVSANAFNGNDD434 pKa = 3.37INLTTTDD441 pKa = 3.41GEE443 pKa = 4.73TYY445 pKa = 10.46ILEE448 pKa = 4.45NYY450 pKa = 8.45TFVAGEE456 pKa = 3.86AKK458 pKa = 9.94FRR460 pKa = 11.84QDD462 pKa = 4.79DD463 pKa = 3.37AWDD466 pKa = 3.9INWGNPAFPTGIATQGGSNITVPAGTFNVFFTRR499 pKa = 11.84STGAYY504 pKa = 8.5EE505 pKa = 4.11FTTVGAFPAVGLIGTAVDD523 pKa = 3.74ANGFDD528 pKa = 5.12GPDD531 pKa = 3.03TDD533 pKa = 3.63MATTNGILYY542 pKa = 8.91TLDD545 pKa = 3.41GVTLMDD551 pKa = 4.16GEE553 pKa = 4.53AKK555 pKa = 10.05FRR557 pKa = 11.84QDD559 pKa = 4.85DD560 pKa = 3.68AWDD563 pKa = 3.84MSWGSVNFPSGTGTVGGANIMVTAGNYY590 pKa = 9.57DD591 pKa = 3.38VTFNRR596 pKa = 11.84LTGDD600 pKa = 3.45YY601 pKa = 10.47SFDD604 pKa = 3.33STLPVYY610 pKa = 10.18PSIGIIGDD618 pKa = 3.53AVTVDD623 pKa = 4.06GFTGADD629 pKa = 2.85VDD631 pKa = 3.94MQTTDD636 pKa = 3.69GITYY640 pKa = 9.28TLMAYY645 pKa = 8.41TFTDD649 pKa = 4.23GEE651 pKa = 4.36AKK653 pKa = 10.01FRR655 pKa = 11.84QDD657 pKa = 5.42DD658 pKa = 3.61NWDD661 pKa = 3.93VNWGSVDD668 pKa = 4.09FPSGTGTQGGSNIEE682 pKa = 4.31VAAGTYY688 pKa = 10.29NVTFNILTGAYY699 pKa = 10.17SFDD702 pKa = 3.53FVNIGILGTALNGFDD717 pKa = 5.56VEE719 pKa = 4.48DD720 pKa = 3.93TDD722 pKa = 3.65MTTTDD727 pKa = 3.19GVNYY731 pKa = 9.87YY732 pKa = 10.74LDD734 pKa = 5.35GIHH737 pKa = 6.41LTDD740 pKa = 3.6GLIKK744 pKa = 10.54FRR746 pKa = 11.84QNDD749 pKa = 3.14NWTVNWGGTDD759 pKa = 3.94FPGGTYY765 pKa = 9.43TLSGEE770 pKa = 4.45NIPVTEE776 pKa = 4.17GDD778 pKa = 3.85YY779 pKa = 10.72IITFNRR785 pKa = 11.84LTGVGQFDD793 pKa = 4.05EE794 pKa = 5.17VILGVNEE801 pKa = 3.79PAQVTFSVFPNPSSEE816 pKa = 3.79SWNFSSRR823 pKa = 11.84GDD825 pKa = 3.62NLHH828 pKa = 7.26DD829 pKa = 4.42IEE831 pKa = 7.03IIDD834 pKa = 3.81ATGKK838 pKa = 8.29TVVSKK843 pKa = 10.92NLSGNHH849 pKa = 4.75ATIDD853 pKa = 3.63ASSLSSGLYY862 pKa = 9.01FAKK865 pKa = 9.52LTSDD869 pKa = 3.92KK870 pKa = 10.85GSKK873 pKa = 8.15TIKK876 pKa = 10.07VIKK879 pKa = 10.1NN880 pKa = 3.24
MM1 pKa = 7.59KK2 pKa = 10.51KK3 pKa = 10.23NLLFLIALICFSLGNAQSISIVGEE27 pKa = 4.0AVGGWEE33 pKa = 4.95DD34 pKa = 4.23NDD36 pKa = 4.09PNTPDD41 pKa = 3.19THH43 pKa = 8.76IMSTVDD49 pKa = 3.31NINYY53 pKa = 7.51TFSNLAVATADD64 pKa = 3.89PNGGGAKK71 pKa = 9.79FRR73 pKa = 11.84QDD75 pKa = 3.37GAWAINWGNAAFPSGTGTQNGANILTVAGTYY106 pKa = 10.55DD107 pKa = 3.26VTFNRR112 pKa = 11.84LTGAYY117 pKa = 9.96SFTNVVTYY125 pKa = 8.94PSIGMIGTAVNANGFNGPDD144 pKa = 3.05VDD146 pKa = 4.19MVTFDD151 pKa = 4.49GITYY155 pKa = 8.38TITNYY160 pKa = 10.43NFLSGEE166 pKa = 4.1MKK168 pKa = 10.44FRR170 pKa = 11.84LDD172 pKa = 4.85DD173 pKa = 3.45SWDD176 pKa = 3.72NSWGSTAFPSGTAVLNVPDD195 pKa = 5.49NIPVPAGTYY204 pKa = 8.9SVSFNRR210 pKa = 11.84ITGDD214 pKa = 3.32YY215 pKa = 10.37SFTFPSIGIIGTAVSAGGFNDD236 pKa = 3.97PDD238 pKa = 4.26TDD240 pKa = 4.4LMTADD245 pKa = 3.34GVHH248 pKa = 5.25YY249 pKa = 10.55VLYY252 pKa = 10.74NHH254 pKa = 6.21VFTDD258 pKa = 4.12GLAKK262 pKa = 10.41FRR264 pKa = 11.84QGDD267 pKa = 3.3NWDD270 pKa = 4.16VNWGNADD277 pKa = 3.25GDD279 pKa = 4.19FPSGIGTQGGTDD291 pKa = 3.18IPVTAGTYY299 pKa = 7.21TVNFNRR305 pKa = 11.84EE306 pKa = 3.37TGAYY310 pKa = 9.95SFDD313 pKa = 3.78APVVFANIGVIGTAVSAGGFDD334 pKa = 5.84DD335 pKa = 5.91PDD337 pKa = 3.58TDD339 pKa = 3.39MTTTDD344 pKa = 4.33GITYY348 pKa = 8.93TLEE351 pKa = 3.95NYY353 pKa = 9.5VFANGQLKK361 pKa = 9.77FRR363 pKa = 11.84QDD365 pKa = 4.12DD366 pKa = 3.56VWDD369 pKa = 4.13NNWGGVGFPGDD380 pKa = 3.37TANFNGPNIPVTAGTYY396 pKa = 9.2TLTFNINTLAYY407 pKa = 9.84SFTGTPLYY415 pKa = 8.21PTVGILGTAVSANAFNGNDD434 pKa = 3.37INLTTTDD441 pKa = 3.41GEE443 pKa = 4.73TYY445 pKa = 10.46ILEE448 pKa = 4.45NYY450 pKa = 8.45TFVAGEE456 pKa = 3.86AKK458 pKa = 9.94FRR460 pKa = 11.84QDD462 pKa = 4.79DD463 pKa = 3.37AWDD466 pKa = 3.9INWGNPAFPTGIATQGGSNITVPAGTFNVFFTRR499 pKa = 11.84STGAYY504 pKa = 8.5EE505 pKa = 4.11FTTVGAFPAVGLIGTAVDD523 pKa = 3.74ANGFDD528 pKa = 5.12GPDD531 pKa = 3.03TDD533 pKa = 3.63MATTNGILYY542 pKa = 8.91TLDD545 pKa = 3.41GVTLMDD551 pKa = 4.16GEE553 pKa = 4.53AKK555 pKa = 10.05FRR557 pKa = 11.84QDD559 pKa = 4.85DD560 pKa = 3.68AWDD563 pKa = 3.84MSWGSVNFPSGTGTVGGANIMVTAGNYY590 pKa = 9.57DD591 pKa = 3.38VTFNRR596 pKa = 11.84LTGDD600 pKa = 3.45YY601 pKa = 10.47SFDD604 pKa = 3.33STLPVYY610 pKa = 10.18PSIGIIGDD618 pKa = 3.53AVTVDD623 pKa = 4.06GFTGADD629 pKa = 2.85VDD631 pKa = 3.94MQTTDD636 pKa = 3.69GITYY640 pKa = 9.28TLMAYY645 pKa = 8.41TFTDD649 pKa = 4.23GEE651 pKa = 4.36AKK653 pKa = 10.01FRR655 pKa = 11.84QDD657 pKa = 5.42DD658 pKa = 3.61NWDD661 pKa = 3.93VNWGSVDD668 pKa = 4.09FPSGTGTQGGSNIEE682 pKa = 4.31VAAGTYY688 pKa = 10.29NVTFNILTGAYY699 pKa = 10.17SFDD702 pKa = 3.53FVNIGILGTALNGFDD717 pKa = 5.56VEE719 pKa = 4.48DD720 pKa = 3.93TDD722 pKa = 3.65MTTTDD727 pKa = 3.19GVNYY731 pKa = 9.87YY732 pKa = 10.74LDD734 pKa = 5.35GIHH737 pKa = 6.41LTDD740 pKa = 3.6GLIKK744 pKa = 10.54FRR746 pKa = 11.84QNDD749 pKa = 3.14NWTVNWGGTDD759 pKa = 3.94FPGGTYY765 pKa = 9.43TLSGEE770 pKa = 4.45NIPVTEE776 pKa = 4.17GDD778 pKa = 3.85YY779 pKa = 10.72IITFNRR785 pKa = 11.84LTGVGQFDD793 pKa = 4.05EE794 pKa = 5.17VILGVNEE801 pKa = 3.79PAQVTFSVFPNPSSEE816 pKa = 3.79SWNFSSRR823 pKa = 11.84GDD825 pKa = 3.62NLHH828 pKa = 7.26DD829 pKa = 4.42IEE831 pKa = 7.03IIDD834 pKa = 3.81ATGKK838 pKa = 8.29TVVSKK843 pKa = 10.92NLSGNHH849 pKa = 4.75ATIDD853 pKa = 3.63ASSLSSGLYY862 pKa = 9.01FAKK865 pKa = 9.52LTSDD869 pKa = 3.92KK870 pKa = 10.85GSKK873 pKa = 8.15TIKK876 pKa = 10.07VIKK879 pKa = 10.1NN880 pKa = 3.24
Molecular weight: 93.56 kDa
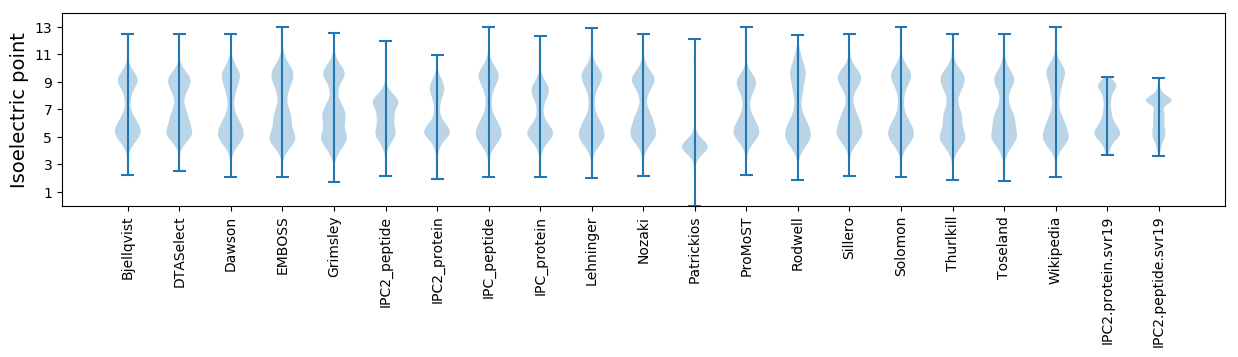
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S1SLR8|A0A2S1SLR8_9FLAO Uncharacterized protein OS=Flavobacterium pallidum OX=2172098 GN=HYN49_12435 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.3RR3 pKa = 11.84RR4 pKa = 11.84IKK6 pKa = 10.44RR7 pKa = 11.84SRR9 pKa = 11.84KK10 pKa = 7.71NSKK13 pKa = 9.39RR14 pKa = 11.84KK15 pKa = 9.44NRR17 pKa = 11.84LGVKK21 pKa = 10.2NSLVNNINARR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.34GVARR38 pKa = 11.84SKK40 pKa = 10.47RR41 pKa = 11.84KK42 pKa = 9.38SHH44 pKa = 6.56ISKK47 pKa = 10.22KK48 pKa = 9.93AYY50 pKa = 9.61KK51 pKa = 10.42AMRR54 pKa = 11.84EE55 pKa = 3.93NWKK58 pKa = 10.31KK59 pKa = 10.89
MM1 pKa = 7.49KK2 pKa = 10.3RR3 pKa = 11.84RR4 pKa = 11.84IKK6 pKa = 10.44RR7 pKa = 11.84SRR9 pKa = 11.84KK10 pKa = 7.71NSKK13 pKa = 9.39RR14 pKa = 11.84KK15 pKa = 9.44NRR17 pKa = 11.84LGVKK21 pKa = 10.2NSLVNNINARR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.34GVARR38 pKa = 11.84SKK40 pKa = 10.47RR41 pKa = 11.84KK42 pKa = 9.38SHH44 pKa = 6.56ISKK47 pKa = 10.22KK48 pKa = 9.93AYY50 pKa = 9.61KK51 pKa = 10.42AMRR54 pKa = 11.84EE55 pKa = 3.93NWKK58 pKa = 10.31KK59 pKa = 10.89
Molecular weight: 7.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1068508 |
22 |
5887 |
364.6 |
40.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.417 ± 0.063 | 0.846 ± 0.02 |
5.748 ± 0.06 | 5.75 ± 0.073 |
5.165 ± 0.045 | 6.876 ± 0.062 |
1.757 ± 0.026 | 7.462 ± 0.05 |
6.86 ± 0.099 | 8.596 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.331 ± 0.03 | 6.133 ± 0.073 |
3.749 ± 0.047 | 3.343 ± 0.028 |
3.395 ± 0.038 | 6.593 ± 0.056 |
6.575 ± 0.139 | 6.38 ± 0.041 |
1.013 ± 0.017 | 4.012 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
