
Pseudaestuariivita atlantica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudaestuariivita
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
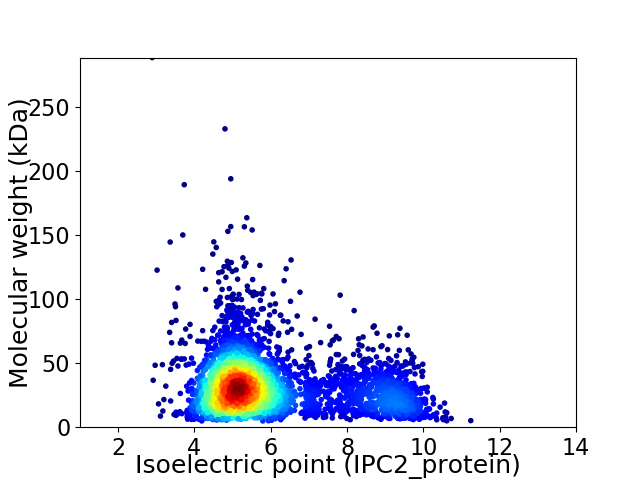
Virtual 2D-PAGE plot for 3978 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
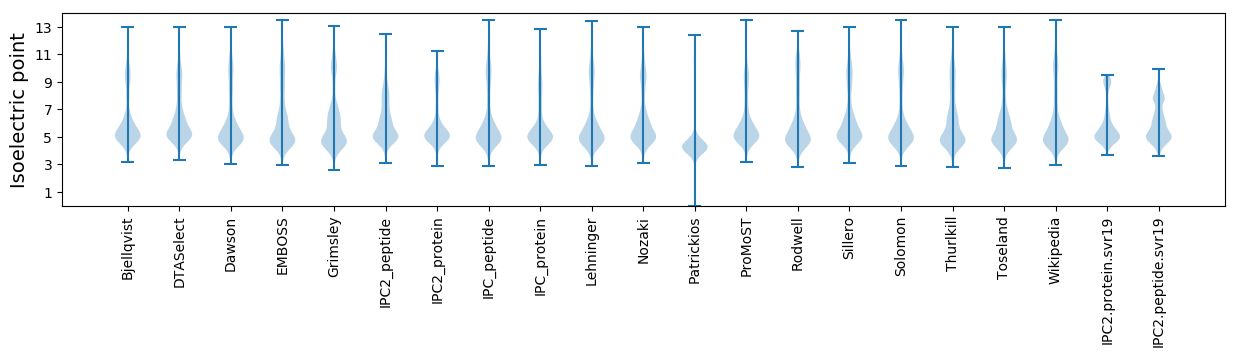
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L1JJ78|A0A0L1JJ78_9RHOB Antirestriction protein OS=Pseudaestuariivita atlantica OX=1317121 GN=ATO11_20670 PE=4 SV=1
MM1 pKa = 7.79AFRR4 pKa = 11.84VWFFVLVGLVFWVPRR19 pKa = 11.84AMAFDD24 pKa = 3.7VTPTFTPTLLQPTQTTRR41 pKa = 11.84LRR43 pKa = 11.84ISFLNSVTSPVTGLTLTDD61 pKa = 3.6TLPAGMFVATPASAVSSCGGSVSTSNGATEE91 pKa = 4.2GTITLTGGTIPAQVGDD107 pKa = 4.01TAGTCYY113 pKa = 10.31IEE115 pKa = 4.3VSVYY119 pKa = 10.02AASSGTYY126 pKa = 9.72INEE129 pKa = 3.82VPANDD134 pKa = 3.4ATGTVAGAPTSNPNPGSATFAATIQNISASMSLDD168 pKa = 2.79IGGNTLQGGEE178 pKa = 4.09TANRR182 pKa = 11.84IITLTNPNDD191 pKa = 3.5VPLTGVSFVHH201 pKa = 7.01DD202 pKa = 4.49LNGDD206 pKa = 3.49SGFNLNANGPVSGTCGGTGTISDD229 pKa = 4.11TTPATSDD236 pKa = 3.23FDD238 pKa = 3.94TTSVITISDD247 pKa = 3.28ATIPANGQCTIIYY260 pKa = 9.26QVEE263 pKa = 4.21PARR266 pKa = 11.84DD267 pKa = 3.43MTRR270 pKa = 11.84TFNDD274 pKa = 2.99TTMTHH279 pKa = 6.3QINGGAITTNEE290 pKa = 4.07GATNPTFNDD299 pKa = 4.53PIRR302 pKa = 11.84TVSGIEE308 pKa = 3.66VDD310 pKa = 3.49KK311 pKa = 11.31TFDD314 pKa = 3.38GVKK317 pKa = 10.21NAEE320 pKa = 4.47LNTNTASGADD330 pKa = 3.67LGIEE334 pKa = 3.74ITNYY338 pKa = 9.9NAAPLPGFTLTDD350 pKa = 3.38TMPAGIVSTNVFRR363 pKa = 11.84NTCGGTSSINGGAEE377 pKa = 3.93VTVTGASLGAASGAPIPGLQVQSCNIDD404 pKa = 3.15ARR406 pKa = 11.84VEE408 pKa = 3.91PSVTGNNTYY417 pKa = 10.64TNVIAAGDD425 pKa = 3.66INGFSHH431 pKa = 7.0TATSANLEE439 pKa = 4.12VTDD442 pKa = 4.45YY443 pKa = 10.93IGPEE447 pKa = 4.17DD448 pKa = 4.09PLSVTKK454 pKa = 10.47AFQMGNIYY462 pKa = 10.44PGDD465 pKa = 3.86TQTLTFTLTNTYY477 pKa = 10.71DD478 pKa = 3.64EE479 pKa = 4.58NLLNVSLTDD488 pKa = 3.41HH489 pKa = 6.74LVNNTSANAGLRR501 pKa = 11.84IGAGGVTSNTCQGSASATPNGDD523 pKa = 4.16LISISGVYY531 pKa = 10.16LPIGASCQVVVEE543 pKa = 4.12VTMASDD549 pKa = 3.58AQNSRR554 pKa = 11.84TIWNTVPQSQITFDD568 pKa = 3.73TVDD571 pKa = 3.27QTGLNPVDD579 pKa = 3.62NAADD583 pKa = 3.65SIYY586 pKa = 10.34VRR588 pKa = 11.84RR589 pKa = 11.84PIEE592 pKa = 4.1LYY594 pKa = 10.48KK595 pKa = 10.87SYY597 pKa = 11.28DD598 pKa = 3.48PASVVSGGEE607 pKa = 3.7SRR609 pKa = 11.84LTLRR613 pKa = 11.84FQRR616 pKa = 11.84NSRR619 pKa = 11.84AGSTTSDD626 pKa = 2.36ITFYY630 pKa = 11.41DD631 pKa = 4.21EE632 pKa = 5.22LPSGHH637 pKa = 6.87TIADD641 pKa = 3.52PANIQNSCGGSVTISPDD658 pKa = 2.97RR659 pKa = 11.84SRR661 pKa = 11.84VDD663 pKa = 3.42LAGGSLGPVGGTTLEE678 pKa = 4.26CTLVLTVIAPNSVGSSNNTVFAGHH702 pKa = 5.62TTATDD707 pKa = 3.75DD708 pKa = 4.14GQPGAFNQMVTTRR721 pKa = 11.84NRR723 pKa = 11.84SATLTRR729 pKa = 11.84TQGDD733 pKa = 3.61LTLTKK738 pKa = 10.5EE739 pKa = 4.25FLQPTIAGGQPSRR752 pKa = 11.84VRR754 pKa = 11.84ITIANTASDD763 pKa = 5.15AIDD766 pKa = 3.55LTGVGITDD774 pKa = 3.96SFAGTEE780 pKa = 3.97LRR782 pKa = 11.84LADD785 pKa = 3.89SVSPTFTNTSGTPNAGGCTGGTFATVGGAGGQITLAGAGVEE826 pKa = 4.23ADD828 pKa = 3.84STCHH832 pKa = 6.72FEE834 pKa = 4.96FNTTSFFGGTVTNSLPAQALVSDD857 pKa = 3.99QGTTNPAPVSASLAVEE873 pKa = 4.12RR874 pKa = 11.84EE875 pKa = 4.17FNVSKK880 pKa = 10.64GFQPAIVGPGEE891 pKa = 3.85TSQAVISVINASSVNFTGTTGIPMLVDD918 pKa = 3.46TLPAGLAISGTPTTTCTGATVSANTASSPQEE949 pKa = 4.0IILSGGSYY957 pKa = 10.11PADD960 pKa = 3.44TSCTIFADD968 pKa = 4.05VSAAAPGAFVNTIPAGSLSTEE989 pKa = 3.96EE990 pKa = 4.62SGTNADD996 pKa = 4.0PASATLAVIAPPTIAKK1012 pKa = 10.26AFGPASIGNGEE1023 pKa = 4.23TSTITFTLSNPNAASLLPAGISGATFSDD1051 pKa = 3.42TMTNMVIAAPPNVGGTCNPQTYY1073 pKa = 9.41SASAGGSVFSVSDD1086 pKa = 3.43LSIAPGASCTVQIDD1100 pKa = 3.72VTSTVPGSHH1109 pKa = 7.51DD1110 pKa = 3.56NQATGISSDD1119 pKa = 3.28QMANPGDD1126 pKa = 3.99PSNIAALTVGTASVPTIAKK1145 pKa = 9.65SFSVDD1150 pKa = 2.89TLLPGEE1156 pKa = 4.27TATLLFTLQNPNAGAANIDD1175 pKa = 3.37AAGFSDD1181 pKa = 4.84TFPTSPGAMVVASPANVYY1199 pKa = 7.44STCDD1203 pKa = 3.16GTVADD1208 pKa = 4.91LGGGSVDD1215 pKa = 4.15PGDD1218 pKa = 4.4TGIQLQGGFIAGNGSCYY1235 pKa = 10.5VSVDD1239 pKa = 3.19VTVPTDD1245 pKa = 3.36GTYY1248 pKa = 10.71TNTSSGLTTEE1258 pKa = 5.17FGTSGTASDD1267 pKa = 3.77TLVAEE1272 pKa = 4.41SAPTILSASEE1282 pKa = 3.82GTRR1285 pKa = 11.84TVRR1288 pKa = 11.84EE1289 pKa = 4.06SDD1291 pKa = 2.95NSQVVDD1297 pKa = 4.1SVLGPNDD1304 pKa = 3.36TFAGQQAFVGSGAAGNVDD1322 pKa = 3.49MTITAIDD1329 pKa = 3.42AALSVNTDD1337 pKa = 2.66TGLIVVAPGTGAGTYY1352 pKa = 10.07SVTYY1356 pKa = 8.13QICQAGNPSNCAPTRR1371 pKa = 11.84TEE1373 pKa = 3.81SVEE1376 pKa = 4.21VVVATILATTDD1387 pKa = 3.64YY1388 pKa = 11.43ASGSPLQVEE1397 pKa = 4.28QADD1400 pKa = 4.1FAKK1403 pKa = 10.8EE1404 pKa = 3.78PGNVLGSSDD1413 pKa = 4.89LLDD1416 pKa = 3.87GVQATVAPGTGGNVDD1431 pKa = 4.23LSILSVTGPSGASTDD1446 pKa = 2.81ITVNVNTGVISVAGSTPQGIYY1467 pKa = 9.87EE1468 pKa = 4.19ISYY1471 pKa = 8.36QICEE1475 pKa = 4.75DD1476 pKa = 3.86DD1477 pKa = 4.85HH1478 pKa = 9.46ADD1480 pKa = 3.69NCDD1483 pKa = 3.34TASEE1487 pKa = 4.3FVEE1490 pKa = 4.37VTIVPIAAGAEE1501 pKa = 4.04TARR1504 pKa = 11.84AATGSDD1510 pKa = 2.87QSVNVGSLLGASDD1523 pKa = 4.08TLDD1526 pKa = 3.71GAPATVGPGATGSVDD1541 pKa = 3.55LSVVSITEE1549 pKa = 4.17GGSPSGNIAVDD1560 pKa = 4.2LDD1562 pKa = 3.74TGDD1565 pKa = 3.64MVVAAGTPDD1574 pKa = 3.14GTYY1577 pKa = 10.42LVTYY1581 pKa = 8.07RR1582 pKa = 11.84ICEE1585 pKa = 3.76EE1586 pKa = 4.27TNRR1589 pKa = 11.84SNCATATEE1597 pKa = 4.91EE1598 pKa = 3.91ITVTSLAIVAGPEE1611 pKa = 3.55ADD1613 pKa = 3.5RR1614 pKa = 11.84TVVGSDD1620 pKa = 3.29GAIDD1624 pKa = 4.19AGPALSSAQDD1634 pKa = 3.33RR1635 pKa = 11.84VDD1637 pKa = 3.78GSAATLGASGNATLSIVPGANTDD1660 pKa = 3.69PEE1662 pKa = 4.49LSIDD1666 pKa = 3.8PVTSRR1671 pKa = 11.84ITVAAGTAAGTYY1683 pKa = 7.98TIEE1686 pKa = 4.45YY1687 pKa = 9.23RR1688 pKa = 11.84LCDD1691 pKa = 3.32STNSDD1696 pKa = 3.13NCATAIEE1703 pKa = 4.38TVIVADD1709 pKa = 4.73RR1710 pKa = 11.84PIAAAPEE1717 pKa = 4.06PVQIVYY1723 pKa = 10.33QSNSAVTAPSIIGASDD1739 pKa = 5.09LIDD1742 pKa = 3.66SAQAVPGFGGNVILTTVSGDD1762 pKa = 3.31PRR1764 pKa = 11.84LTLDD1768 pKa = 3.56TEE1770 pKa = 4.23TGEE1773 pKa = 4.54IEE1775 pKa = 4.47VAPNMPLGDD1784 pKa = 3.56FTLTYY1789 pKa = 10.0RR1790 pKa = 11.84ICDD1793 pKa = 3.52PANLDD1798 pKa = 3.44NCATATEE1805 pKa = 4.48TVRR1808 pKa = 11.84IARR1811 pKa = 11.84RR1812 pKa = 11.84PITAASEE1819 pKa = 4.12ADD1821 pKa = 3.62RR1822 pKa = 11.84PLYY1825 pKa = 10.78ASTAAQTAGSVIGSGDD1841 pKa = 3.64TLDD1844 pKa = 4.5GLAAVPGFGGNVVLTLGGGSPEE1866 pKa = 3.84VSLNVATGDD1875 pKa = 3.7LGVAAGTPPGTYY1887 pKa = 10.43VLDD1890 pKa = 4.1YY1891 pKa = 11.2NN1892 pKa = 4.57
MM1 pKa = 7.79AFRR4 pKa = 11.84VWFFVLVGLVFWVPRR19 pKa = 11.84AMAFDD24 pKa = 3.7VTPTFTPTLLQPTQTTRR41 pKa = 11.84LRR43 pKa = 11.84ISFLNSVTSPVTGLTLTDD61 pKa = 3.6TLPAGMFVATPASAVSSCGGSVSTSNGATEE91 pKa = 4.2GTITLTGGTIPAQVGDD107 pKa = 4.01TAGTCYY113 pKa = 10.31IEE115 pKa = 4.3VSVYY119 pKa = 10.02AASSGTYY126 pKa = 9.72INEE129 pKa = 3.82VPANDD134 pKa = 3.4ATGTVAGAPTSNPNPGSATFAATIQNISASMSLDD168 pKa = 2.79IGGNTLQGGEE178 pKa = 4.09TANRR182 pKa = 11.84IITLTNPNDD191 pKa = 3.5VPLTGVSFVHH201 pKa = 7.01DD202 pKa = 4.49LNGDD206 pKa = 3.49SGFNLNANGPVSGTCGGTGTISDD229 pKa = 4.11TTPATSDD236 pKa = 3.23FDD238 pKa = 3.94TTSVITISDD247 pKa = 3.28ATIPANGQCTIIYY260 pKa = 9.26QVEE263 pKa = 4.21PARR266 pKa = 11.84DD267 pKa = 3.43MTRR270 pKa = 11.84TFNDD274 pKa = 2.99TTMTHH279 pKa = 6.3QINGGAITTNEE290 pKa = 4.07GATNPTFNDD299 pKa = 4.53PIRR302 pKa = 11.84TVSGIEE308 pKa = 3.66VDD310 pKa = 3.49KK311 pKa = 11.31TFDD314 pKa = 3.38GVKK317 pKa = 10.21NAEE320 pKa = 4.47LNTNTASGADD330 pKa = 3.67LGIEE334 pKa = 3.74ITNYY338 pKa = 9.9NAAPLPGFTLTDD350 pKa = 3.38TMPAGIVSTNVFRR363 pKa = 11.84NTCGGTSSINGGAEE377 pKa = 3.93VTVTGASLGAASGAPIPGLQVQSCNIDD404 pKa = 3.15ARR406 pKa = 11.84VEE408 pKa = 3.91PSVTGNNTYY417 pKa = 10.64TNVIAAGDD425 pKa = 3.66INGFSHH431 pKa = 7.0TATSANLEE439 pKa = 4.12VTDD442 pKa = 4.45YY443 pKa = 10.93IGPEE447 pKa = 4.17DD448 pKa = 4.09PLSVTKK454 pKa = 10.47AFQMGNIYY462 pKa = 10.44PGDD465 pKa = 3.86TQTLTFTLTNTYY477 pKa = 10.71DD478 pKa = 3.64EE479 pKa = 4.58NLLNVSLTDD488 pKa = 3.41HH489 pKa = 6.74LVNNTSANAGLRR501 pKa = 11.84IGAGGVTSNTCQGSASATPNGDD523 pKa = 4.16LISISGVYY531 pKa = 10.16LPIGASCQVVVEE543 pKa = 4.12VTMASDD549 pKa = 3.58AQNSRR554 pKa = 11.84TIWNTVPQSQITFDD568 pKa = 3.73TVDD571 pKa = 3.27QTGLNPVDD579 pKa = 3.62NAADD583 pKa = 3.65SIYY586 pKa = 10.34VRR588 pKa = 11.84RR589 pKa = 11.84PIEE592 pKa = 4.1LYY594 pKa = 10.48KK595 pKa = 10.87SYY597 pKa = 11.28DD598 pKa = 3.48PASVVSGGEE607 pKa = 3.7SRR609 pKa = 11.84LTLRR613 pKa = 11.84FQRR616 pKa = 11.84NSRR619 pKa = 11.84AGSTTSDD626 pKa = 2.36ITFYY630 pKa = 11.41DD631 pKa = 4.21EE632 pKa = 5.22LPSGHH637 pKa = 6.87TIADD641 pKa = 3.52PANIQNSCGGSVTISPDD658 pKa = 2.97RR659 pKa = 11.84SRR661 pKa = 11.84VDD663 pKa = 3.42LAGGSLGPVGGTTLEE678 pKa = 4.26CTLVLTVIAPNSVGSSNNTVFAGHH702 pKa = 5.62TTATDD707 pKa = 3.75DD708 pKa = 4.14GQPGAFNQMVTTRR721 pKa = 11.84NRR723 pKa = 11.84SATLTRR729 pKa = 11.84TQGDD733 pKa = 3.61LTLTKK738 pKa = 10.5EE739 pKa = 4.25FLQPTIAGGQPSRR752 pKa = 11.84VRR754 pKa = 11.84ITIANTASDD763 pKa = 5.15AIDD766 pKa = 3.55LTGVGITDD774 pKa = 3.96SFAGTEE780 pKa = 3.97LRR782 pKa = 11.84LADD785 pKa = 3.89SVSPTFTNTSGTPNAGGCTGGTFATVGGAGGQITLAGAGVEE826 pKa = 4.23ADD828 pKa = 3.84STCHH832 pKa = 6.72FEE834 pKa = 4.96FNTTSFFGGTVTNSLPAQALVSDD857 pKa = 3.99QGTTNPAPVSASLAVEE873 pKa = 4.12RR874 pKa = 11.84EE875 pKa = 4.17FNVSKK880 pKa = 10.64GFQPAIVGPGEE891 pKa = 3.85TSQAVISVINASSVNFTGTTGIPMLVDD918 pKa = 3.46TLPAGLAISGTPTTTCTGATVSANTASSPQEE949 pKa = 4.0IILSGGSYY957 pKa = 10.11PADD960 pKa = 3.44TSCTIFADD968 pKa = 4.05VSAAAPGAFVNTIPAGSLSTEE989 pKa = 3.96EE990 pKa = 4.62SGTNADD996 pKa = 4.0PASATLAVIAPPTIAKK1012 pKa = 10.26AFGPASIGNGEE1023 pKa = 4.23TSTITFTLSNPNAASLLPAGISGATFSDD1051 pKa = 3.42TMTNMVIAAPPNVGGTCNPQTYY1073 pKa = 9.41SASAGGSVFSVSDD1086 pKa = 3.43LSIAPGASCTVQIDD1100 pKa = 3.72VTSTVPGSHH1109 pKa = 7.51DD1110 pKa = 3.56NQATGISSDD1119 pKa = 3.28QMANPGDD1126 pKa = 3.99PSNIAALTVGTASVPTIAKK1145 pKa = 9.65SFSVDD1150 pKa = 2.89TLLPGEE1156 pKa = 4.27TATLLFTLQNPNAGAANIDD1175 pKa = 3.37AAGFSDD1181 pKa = 4.84TFPTSPGAMVVASPANVYY1199 pKa = 7.44STCDD1203 pKa = 3.16GTVADD1208 pKa = 4.91LGGGSVDD1215 pKa = 4.15PGDD1218 pKa = 4.4TGIQLQGGFIAGNGSCYY1235 pKa = 10.5VSVDD1239 pKa = 3.19VTVPTDD1245 pKa = 3.36GTYY1248 pKa = 10.71TNTSSGLTTEE1258 pKa = 5.17FGTSGTASDD1267 pKa = 3.77TLVAEE1272 pKa = 4.41SAPTILSASEE1282 pKa = 3.82GTRR1285 pKa = 11.84TVRR1288 pKa = 11.84EE1289 pKa = 4.06SDD1291 pKa = 2.95NSQVVDD1297 pKa = 4.1SVLGPNDD1304 pKa = 3.36TFAGQQAFVGSGAAGNVDD1322 pKa = 3.49MTITAIDD1329 pKa = 3.42AALSVNTDD1337 pKa = 2.66TGLIVVAPGTGAGTYY1352 pKa = 10.07SVTYY1356 pKa = 8.13QICQAGNPSNCAPTRR1371 pKa = 11.84TEE1373 pKa = 3.81SVEE1376 pKa = 4.21VVVATILATTDD1387 pKa = 3.64YY1388 pKa = 11.43ASGSPLQVEE1397 pKa = 4.28QADD1400 pKa = 4.1FAKK1403 pKa = 10.8EE1404 pKa = 3.78PGNVLGSSDD1413 pKa = 4.89LLDD1416 pKa = 3.87GVQATVAPGTGGNVDD1431 pKa = 4.23LSILSVTGPSGASTDD1446 pKa = 2.81ITVNVNTGVISVAGSTPQGIYY1467 pKa = 9.87EE1468 pKa = 4.19ISYY1471 pKa = 8.36QICEE1475 pKa = 4.75DD1476 pKa = 3.86DD1477 pKa = 4.85HH1478 pKa = 9.46ADD1480 pKa = 3.69NCDD1483 pKa = 3.34TASEE1487 pKa = 4.3FVEE1490 pKa = 4.37VTIVPIAAGAEE1501 pKa = 4.04TARR1504 pKa = 11.84AATGSDD1510 pKa = 2.87QSVNVGSLLGASDD1523 pKa = 4.08TLDD1526 pKa = 3.71GAPATVGPGATGSVDD1541 pKa = 3.55LSVVSITEE1549 pKa = 4.17GGSPSGNIAVDD1560 pKa = 4.2LDD1562 pKa = 3.74TGDD1565 pKa = 3.64MVVAAGTPDD1574 pKa = 3.14GTYY1577 pKa = 10.42LVTYY1581 pKa = 8.07RR1582 pKa = 11.84ICEE1585 pKa = 3.76EE1586 pKa = 4.27TNRR1589 pKa = 11.84SNCATATEE1597 pKa = 4.91EE1598 pKa = 3.91ITVTSLAIVAGPEE1611 pKa = 3.55ADD1613 pKa = 3.5RR1614 pKa = 11.84TVVGSDD1620 pKa = 3.29GAIDD1624 pKa = 4.19AGPALSSAQDD1634 pKa = 3.33RR1635 pKa = 11.84VDD1637 pKa = 3.78GSAATLGASGNATLSIVPGANTDD1660 pKa = 3.69PEE1662 pKa = 4.49LSIDD1666 pKa = 3.8PVTSRR1671 pKa = 11.84ITVAAGTAAGTYY1683 pKa = 7.98TIEE1686 pKa = 4.45YY1687 pKa = 9.23RR1688 pKa = 11.84LCDD1691 pKa = 3.32STNSDD1696 pKa = 3.13NCATAIEE1703 pKa = 4.38TVIVADD1709 pKa = 4.73RR1710 pKa = 11.84PIAAAPEE1717 pKa = 4.06PVQIVYY1723 pKa = 10.33QSNSAVTAPSIIGASDD1739 pKa = 5.09LIDD1742 pKa = 3.66SAQAVPGFGGNVILTTVSGDD1762 pKa = 3.31PRR1764 pKa = 11.84LTLDD1768 pKa = 3.56TEE1770 pKa = 4.23TGEE1773 pKa = 4.54IEE1775 pKa = 4.47VAPNMPLGDD1784 pKa = 3.56FTLTYY1789 pKa = 10.0RR1790 pKa = 11.84ICDD1793 pKa = 3.52PANLDD1798 pKa = 3.44NCATATEE1805 pKa = 4.48TVRR1808 pKa = 11.84IARR1811 pKa = 11.84RR1812 pKa = 11.84PITAASEE1819 pKa = 4.12ADD1821 pKa = 3.62RR1822 pKa = 11.84PLYY1825 pKa = 10.78ASTAAQTAGSVIGSGDD1841 pKa = 3.64TLDD1844 pKa = 4.5GLAAVPGFGGNVVLTLGGGSPEE1866 pKa = 3.84VSLNVATGDD1875 pKa = 3.7LGVAAGTPPGTYY1887 pKa = 10.43VLDD1890 pKa = 4.1YY1891 pKa = 11.2NN1892 pKa = 4.57
Molecular weight: 189.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L1JKN8|A0A0L1JKN8_9RHOB Cytochrome B OS=Pseudaestuariivita atlantica OX=1317121 GN=ATO11_17005 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1234050 |
41 |
2946 |
310.2 |
33.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.817 ± 0.05 | 0.897 ± 0.014 |
6.561 ± 0.043 | 5.672 ± 0.038 |
3.706 ± 0.023 | 8.857 ± 0.047 |
2.082 ± 0.02 | 4.905 ± 0.025 |
2.817 ± 0.034 | 9.824 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.753 ± 0.021 | 2.412 ± 0.024 |
5.172 ± 0.034 | 2.969 ± 0.02 |
6.94 ± 0.039 | 4.747 ± 0.022 |
5.69 ± 0.033 | 7.522 ± 0.034 |
1.474 ± 0.017 | 2.182 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
