
Psychroserpens burtonensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Psychroserpens
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
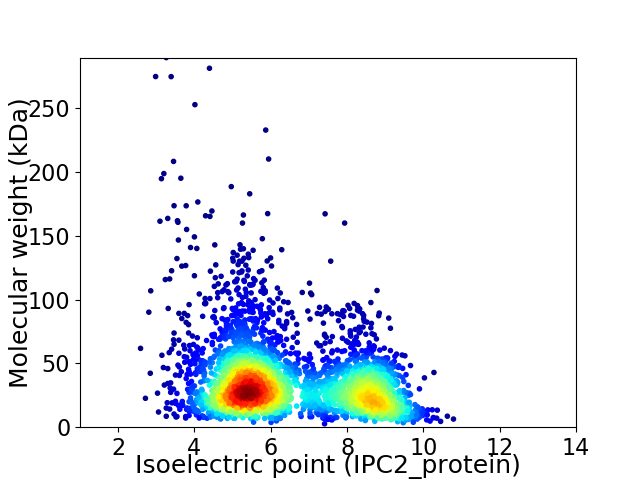
Virtual 2D-PAGE plot for 3458 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
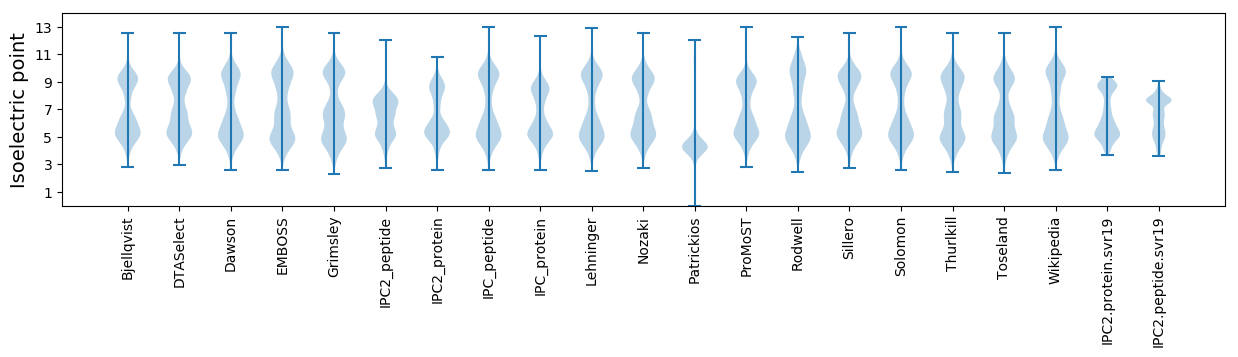
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C7B7S8|A0A5C7B7S8_9FLAO Sigma-70 family RNA polymerase sigma factor OS=Psychroserpens burtonensis OX=49278 GN=ES692_14790 PE=3 SV=1
MM1 pKa = 7.66KK2 pKa = 10.49NLIYY6 pKa = 10.31IIAILLTVGMFQSCSDD22 pKa = 4.13DD23 pKa = 4.23DD24 pKa = 4.88FPVPPASTVPEE35 pKa = 4.02FAFVIDD41 pKa = 3.8NDD43 pKa = 3.62EE44 pKa = 4.24FAPATVTFTNNSIVPGNVGTATYY67 pKa = 9.9YY68 pKa = 10.31WNFGDD73 pKa = 4.97GEE75 pKa = 4.6STSEE79 pKa = 3.98EE80 pKa = 4.26NPTYY84 pKa = 10.78LYY86 pKa = 9.99TEE88 pKa = 4.21SGAYY92 pKa = 8.01TVNLVVTTSEE102 pKa = 3.9SLEE105 pKa = 3.86INEE108 pKa = 3.85ITKK111 pKa = 9.22TVVIKK116 pKa = 10.64DD117 pKa = 3.66PNATGVPIYY126 pKa = 9.34FTDD129 pKa = 4.62GAQVYY134 pKa = 9.85QGLINTQAPIFTTVNGVVAQSSYY157 pKa = 11.54GMVIDD162 pKa = 4.09NVNSKK167 pKa = 10.86LYY169 pKa = 10.35ISDD172 pKa = 3.62YY173 pKa = 10.96GADD176 pKa = 3.3KK177 pKa = 10.41IYY179 pKa = 10.65RR180 pKa = 11.84SNLDD184 pKa = 2.95GSNFEE189 pKa = 4.16EE190 pKa = 4.73FRR192 pKa = 11.84TGVDD196 pKa = 3.24QPNGMTIDD204 pKa = 3.66YY205 pKa = 10.0EE206 pKa = 4.28EE207 pKa = 4.14NQIYY211 pKa = 9.86WDD213 pKa = 3.86TSSGIQRR220 pKa = 11.84GNIDD224 pKa = 3.89DD225 pKa = 3.98TDD227 pKa = 3.83VNQVEE232 pKa = 4.79DD233 pKa = 4.32FVTGQSNDD241 pKa = 3.52PDD243 pKa = 4.12GLSIDD248 pKa = 3.72LVNRR252 pKa = 11.84KK253 pKa = 9.55LYY255 pKa = 7.87WVNYY259 pKa = 9.37NGGVWVKK266 pKa = 10.6NLDD269 pKa = 3.65GTGEE273 pKa = 4.15SEE275 pKa = 5.04LIPVIEE281 pKa = 4.75GGSMLVIGDD290 pKa = 4.3RR291 pKa = 11.84IFYY294 pKa = 10.41DD295 pKa = 3.78EE296 pKa = 4.53YY297 pKa = 11.0VASGDD302 pKa = 3.72VRR304 pKa = 11.84LKK306 pKa = 10.96SADD309 pKa = 3.71LDD311 pKa = 3.72GTNISTIAVNIGRR324 pKa = 11.84VVYY327 pKa = 10.28GIGYY331 pKa = 8.29DD332 pKa = 3.4ANEE335 pKa = 3.95NKK337 pKa = 10.01IYY339 pKa = 10.15WGDD342 pKa = 3.52RR343 pKa = 11.84STDD346 pKa = 2.83VMMRR350 pKa = 11.84ANLDD354 pKa = 3.46GSSPEE359 pKa = 3.43AWFTHH364 pKa = 5.82SADD367 pKa = 3.06TRR369 pKa = 11.84GIVIGEE375 pKa = 4.22LEE377 pKa = 3.88
MM1 pKa = 7.66KK2 pKa = 10.49NLIYY6 pKa = 10.31IIAILLTVGMFQSCSDD22 pKa = 4.13DD23 pKa = 4.23DD24 pKa = 4.88FPVPPASTVPEE35 pKa = 4.02FAFVIDD41 pKa = 3.8NDD43 pKa = 3.62EE44 pKa = 4.24FAPATVTFTNNSIVPGNVGTATYY67 pKa = 9.9YY68 pKa = 10.31WNFGDD73 pKa = 4.97GEE75 pKa = 4.6STSEE79 pKa = 3.98EE80 pKa = 4.26NPTYY84 pKa = 10.78LYY86 pKa = 9.99TEE88 pKa = 4.21SGAYY92 pKa = 8.01TVNLVVTTSEE102 pKa = 3.9SLEE105 pKa = 3.86INEE108 pKa = 3.85ITKK111 pKa = 9.22TVVIKK116 pKa = 10.64DD117 pKa = 3.66PNATGVPIYY126 pKa = 9.34FTDD129 pKa = 4.62GAQVYY134 pKa = 9.85QGLINTQAPIFTTVNGVVAQSSYY157 pKa = 11.54GMVIDD162 pKa = 4.09NVNSKK167 pKa = 10.86LYY169 pKa = 10.35ISDD172 pKa = 3.62YY173 pKa = 10.96GADD176 pKa = 3.3KK177 pKa = 10.41IYY179 pKa = 10.65RR180 pKa = 11.84SNLDD184 pKa = 2.95GSNFEE189 pKa = 4.16EE190 pKa = 4.73FRR192 pKa = 11.84TGVDD196 pKa = 3.24QPNGMTIDD204 pKa = 3.66YY205 pKa = 10.0EE206 pKa = 4.28EE207 pKa = 4.14NQIYY211 pKa = 9.86WDD213 pKa = 3.86TSSGIQRR220 pKa = 11.84GNIDD224 pKa = 3.89DD225 pKa = 3.98TDD227 pKa = 3.83VNQVEE232 pKa = 4.79DD233 pKa = 4.32FVTGQSNDD241 pKa = 3.52PDD243 pKa = 4.12GLSIDD248 pKa = 3.72LVNRR252 pKa = 11.84KK253 pKa = 9.55LYY255 pKa = 7.87WVNYY259 pKa = 9.37NGGVWVKK266 pKa = 10.6NLDD269 pKa = 3.65GTGEE273 pKa = 4.15SEE275 pKa = 5.04LIPVIEE281 pKa = 4.75GGSMLVIGDD290 pKa = 4.3RR291 pKa = 11.84IFYY294 pKa = 10.41DD295 pKa = 3.78EE296 pKa = 4.53YY297 pKa = 11.0VASGDD302 pKa = 3.72VRR304 pKa = 11.84LKK306 pKa = 10.96SADD309 pKa = 3.71LDD311 pKa = 3.72GTNISTIAVNIGRR324 pKa = 11.84VVYY327 pKa = 10.28GIGYY331 pKa = 8.29DD332 pKa = 3.4ANEE335 pKa = 3.95NKK337 pKa = 10.01IYY339 pKa = 10.15WGDD342 pKa = 3.52RR343 pKa = 11.84STDD346 pKa = 2.83VMMRR350 pKa = 11.84ANLDD354 pKa = 3.46GSSPEE359 pKa = 3.43AWFTHH364 pKa = 5.82SADD367 pKa = 3.06TRR369 pKa = 11.84GIVIGEE375 pKa = 4.22LEE377 pKa = 3.88
Molecular weight: 41.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C7BE50|A0A5C7BE50_9FLAO Uncharacterized protein OS=Psychroserpens burtonensis OX=49278 GN=ES692_09620 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.04TRR50 pKa = 11.84HH51 pKa = 5.93KK52 pKa = 10.8RR53 pKa = 3.27
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.04TRR50 pKa = 11.84HH51 pKa = 5.93KK52 pKa = 10.8RR53 pKa = 3.27
Molecular weight: 6.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1151945 |
22 |
2740 |
333.1 |
37.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.119 ± 0.043 | 0.773 ± 0.016 |
5.985 ± 0.039 | 6.314 ± 0.041 |
5.27 ± 0.041 | 6.233 ± 0.045 |
1.752 ± 0.023 | 8.289 ± 0.039 |
7.53 ± 0.065 | 9.249 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.021 | 6.394 ± 0.045 |
3.172 ± 0.02 | 3.375 ± 0.023 |
3.301 ± 0.029 | 6.76 ± 0.033 |
6.205 ± 0.053 | 6.138 ± 0.034 |
0.99 ± 0.015 | 4.018 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
