
Sphaerochaeta coccoides (strain ATCC BAA-1237 / DSM 17374 / SPN1) (Spirochaeta coccoides)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Sphaerochaetaceae; Sphaerochaeta; Sphaerochaeta coccoides
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
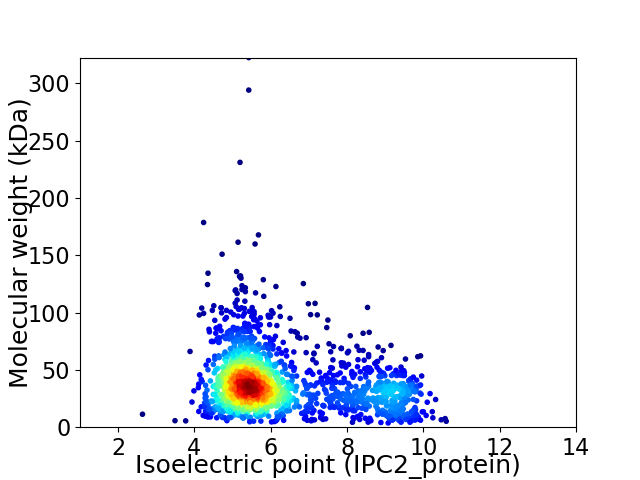
Virtual 2D-PAGE plot for 1819 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4GM11|F4GM11_SPHCD Uncharacterized protein OS=Sphaerochaeta coccoides (strain ATCC BAA-1237 / DSM 17374 / SPN1) OX=760011 GN=Spico_1278 PE=4 SV=1
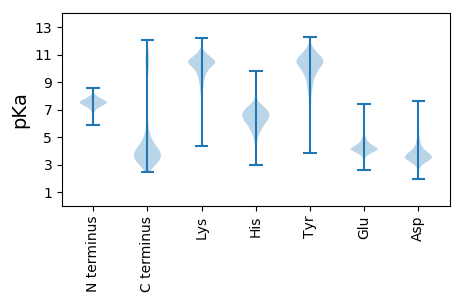
MM1 pKa = 7.72KK2 pKa = 10.32KK3 pKa = 10.09RR4 pKa = 11.84LFVLMVFLGVSCVVLFAGPFGIDD27 pKa = 2.71MKK29 pKa = 10.82MSLEE33 pKa = 4.09DD34 pKa = 3.76LEE36 pKa = 4.54NAGFFPEE43 pKa = 3.78VDD45 pKa = 4.24EE46 pKa = 4.83IRR48 pKa = 11.84TVYY51 pKa = 8.52TWYY54 pKa = 10.84SVFPSKK60 pKa = 10.57PHH62 pKa = 7.12DD63 pKa = 3.78LFEE66 pKa = 4.54QYY68 pKa = 9.92FVAVDD73 pKa = 3.84DD74 pKa = 4.85EE75 pKa = 4.87YY76 pKa = 11.77GVCKK80 pKa = 10.31IVAVGKK86 pKa = 10.13DD87 pKa = 3.54IEE89 pKa = 4.42TSTYY93 pKa = 9.62GEE95 pKa = 4.01NVIDD99 pKa = 3.67AFEE102 pKa = 4.02RR103 pKa = 11.84VEE105 pKa = 5.15KK106 pKa = 10.37GVSWTYY112 pKa = 11.48GEE114 pKa = 4.02VDD116 pKa = 3.66VVFDD120 pKa = 3.8YY121 pKa = 11.59LDD123 pKa = 3.97YY124 pKa = 11.04EE125 pKa = 4.87SVWNEE130 pKa = 3.82PEE132 pKa = 3.38DD133 pKa = 3.53WMYY136 pKa = 11.49GLYY139 pKa = 10.27YY140 pKa = 10.08EE141 pKa = 4.92EE142 pKa = 4.1RR143 pKa = 11.84TLLAFWEE150 pKa = 4.4PTLDD154 pKa = 5.12DD155 pKa = 4.22IDD157 pKa = 6.36NIMLEE162 pKa = 4.27ANALSSTVGWLMLGYY177 pKa = 10.46EE178 pKa = 4.27SLHH181 pKa = 6.1WSDD184 pKa = 4.53VLDD187 pKa = 4.42RR188 pKa = 11.84DD189 pKa = 3.73
MM1 pKa = 7.72KK2 pKa = 10.32KK3 pKa = 10.09RR4 pKa = 11.84LFVLMVFLGVSCVVLFAGPFGIDD27 pKa = 2.71MKK29 pKa = 10.82MSLEE33 pKa = 4.09DD34 pKa = 3.76LEE36 pKa = 4.54NAGFFPEE43 pKa = 3.78VDD45 pKa = 4.24EE46 pKa = 4.83IRR48 pKa = 11.84TVYY51 pKa = 8.52TWYY54 pKa = 10.84SVFPSKK60 pKa = 10.57PHH62 pKa = 7.12DD63 pKa = 3.78LFEE66 pKa = 4.54QYY68 pKa = 9.92FVAVDD73 pKa = 3.84DD74 pKa = 4.85EE75 pKa = 4.87YY76 pKa = 11.77GVCKK80 pKa = 10.31IVAVGKK86 pKa = 10.13DD87 pKa = 3.54IEE89 pKa = 4.42TSTYY93 pKa = 9.62GEE95 pKa = 4.01NVIDD99 pKa = 3.67AFEE102 pKa = 4.02RR103 pKa = 11.84VEE105 pKa = 5.15KK106 pKa = 10.37GVSWTYY112 pKa = 11.48GEE114 pKa = 4.02VDD116 pKa = 3.66VVFDD120 pKa = 3.8YY121 pKa = 11.59LDD123 pKa = 3.97YY124 pKa = 11.04EE125 pKa = 4.87SVWNEE130 pKa = 3.82PEE132 pKa = 3.38DD133 pKa = 3.53WMYY136 pKa = 11.49GLYY139 pKa = 10.27YY140 pKa = 10.08EE141 pKa = 4.92EE142 pKa = 4.1RR143 pKa = 11.84TLLAFWEE150 pKa = 4.4PTLDD154 pKa = 5.12DD155 pKa = 4.22IDD157 pKa = 6.36NIMLEE162 pKa = 4.27ANALSSTVGWLMLGYY177 pKa = 10.46EE178 pKa = 4.27SLHH181 pKa = 6.1WSDD184 pKa = 4.53VLDD187 pKa = 4.42RR188 pKa = 11.84DD189 pKa = 3.73
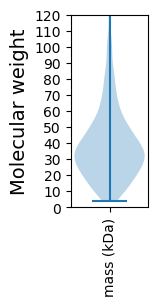
Molecular weight: 21.96 kDa
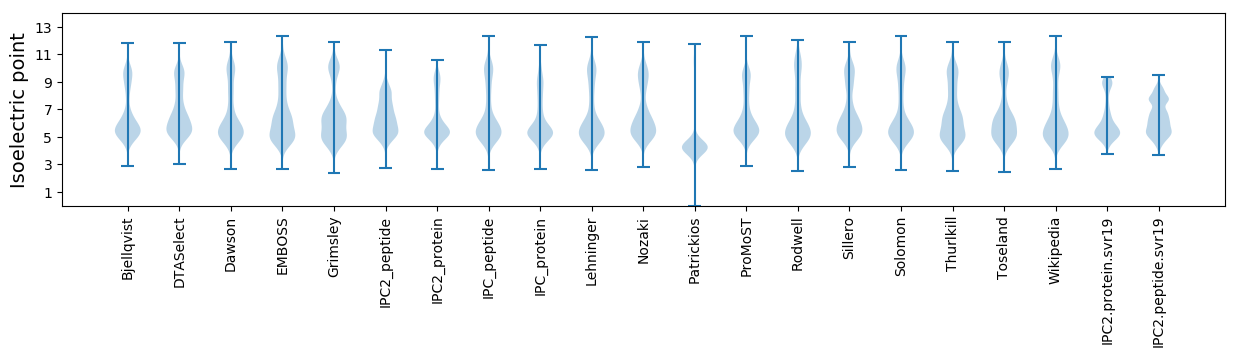
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4GLS9|F4GLS9_SPHCD Uncharacterized protein OS=Sphaerochaeta coccoides (strain ATCC BAA-1237 / DSM 17374 / SPN1) OX=760011 GN=Spico_1264 PE=4 SV=1
MM1 pKa = 7.56NKK3 pKa = 9.57TIQTTLKK10 pKa = 10.35RR11 pKa = 11.84MSGRR15 pKa = 11.84NEE17 pKa = 3.72PYY19 pKa = 10.09IFLILILLSILIQVRR34 pKa = 11.84SGQFFSSNNLVDD46 pKa = 3.29IASALIVPGLFAVATFMVIVSGGIDD71 pKa = 3.28VSFPALASLSVYY83 pKa = 10.59AITKK87 pKa = 7.87MFLDD91 pKa = 3.88IGYY94 pKa = 9.33QGNVLLPLACAVFLGALLGAFNGLFVGYY122 pKa = 9.79FGLPAMIVTLGSSSVFRR139 pKa = 11.84GIMQGALNSRR149 pKa = 11.84QLTMIPEE156 pKa = 4.41PMRR159 pKa = 11.84IFGTTPLFIARR170 pKa = 11.84NPVSGLTSRR179 pKa = 11.84LPLPFMILVVVAVLVFLMMRR199 pKa = 11.84FTMFGRR205 pKa = 11.84GIYY208 pKa = 10.22AIGGSEE214 pKa = 4.17VSAHH218 pKa = 6.08RR219 pKa = 11.84AGFHH223 pKa = 5.13VKK225 pKa = 8.36RR226 pKa = 11.84TKK228 pKa = 10.39FLLYY232 pKa = 10.25VFAGIIASLAGFARR246 pKa = 11.84VTMMQQAHH254 pKa = 5.98PTNMLGMEE262 pKa = 4.23MNIIAGVVLGGTAITGGRR280 pKa = 11.84GTLLGCFLGTLLIVIVEE297 pKa = 4.1NSLILIGVPTSWKK310 pKa = 10.47SVFTGLLIIIGTGVSAYY327 pKa = 10.71QGMLMGKK334 pKa = 9.45KK335 pKa = 9.84RR336 pKa = 11.84GIGTKK341 pKa = 7.07EE342 pKa = 3.65VRR344 pKa = 11.84SS345 pKa = 3.82
MM1 pKa = 7.56NKK3 pKa = 9.57TIQTTLKK10 pKa = 10.35RR11 pKa = 11.84MSGRR15 pKa = 11.84NEE17 pKa = 3.72PYY19 pKa = 10.09IFLILILLSILIQVRR34 pKa = 11.84SGQFFSSNNLVDD46 pKa = 3.29IASALIVPGLFAVATFMVIVSGGIDD71 pKa = 3.28VSFPALASLSVYY83 pKa = 10.59AITKK87 pKa = 7.87MFLDD91 pKa = 3.88IGYY94 pKa = 9.33QGNVLLPLACAVFLGALLGAFNGLFVGYY122 pKa = 9.79FGLPAMIVTLGSSSVFRR139 pKa = 11.84GIMQGALNSRR149 pKa = 11.84QLTMIPEE156 pKa = 4.41PMRR159 pKa = 11.84IFGTTPLFIARR170 pKa = 11.84NPVSGLTSRR179 pKa = 11.84LPLPFMILVVVAVLVFLMMRR199 pKa = 11.84FTMFGRR205 pKa = 11.84GIYY208 pKa = 10.22AIGGSEE214 pKa = 4.17VSAHH218 pKa = 6.08RR219 pKa = 11.84AGFHH223 pKa = 5.13VKK225 pKa = 8.36RR226 pKa = 11.84TKK228 pKa = 10.39FLLYY232 pKa = 10.25VFAGIIASLAGFARR246 pKa = 11.84VTMMQQAHH254 pKa = 5.98PTNMLGMEE262 pKa = 4.23MNIIAGVVLGGTAITGGRR280 pKa = 11.84GTLLGCFLGTLLIVIVEE297 pKa = 4.1NSLILIGVPTSWKK310 pKa = 10.47SVFTGLLIIIGTGVSAYY327 pKa = 10.71QGMLMGKK334 pKa = 9.45KK335 pKa = 9.84RR336 pKa = 11.84GIGTKK341 pKa = 7.07EE342 pKa = 3.65VRR344 pKa = 11.84SS345 pKa = 3.82
Molecular weight: 36.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
660275 |
34 |
2867 |
363.0 |
40.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.477 ± 0.052 | 1.022 ± 0.019 |
5.842 ± 0.044 | 5.91 ± 0.06 |
4.335 ± 0.045 | 7.859 ± 0.059 |
2.101 ± 0.023 | 6.672 ± 0.049 |
4.969 ± 0.049 | 9.41 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.909 ± 0.027 | 3.573 ± 0.038 |
4.101 ± 0.035 | 2.939 ± 0.032 |
5.379 ± 0.046 | 6.864 ± 0.049 |
5.935 ± 0.057 | 7.291 ± 0.045 |
1.195 ± 0.021 | 3.216 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
