
Nocardioides sp. Root1257
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
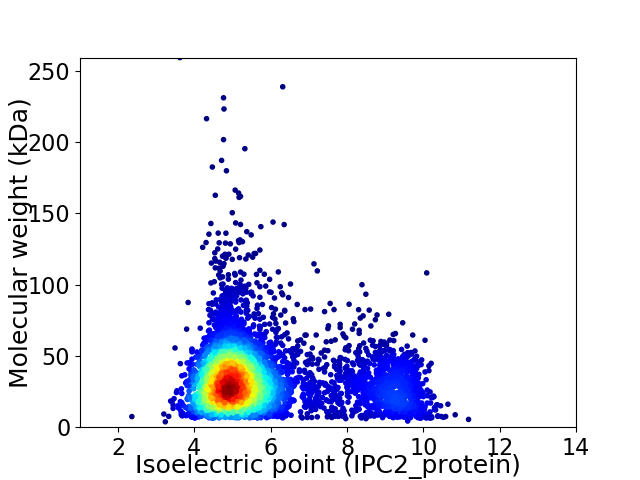
Virtual 2D-PAGE plot for 5063 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7BHV4|A0A0Q7BHV4_9ACTN Chaperone protein DnaJ OS=Nocardioides sp. Root1257 OX=1736439 GN=dnaJ PE=3 SV=1
MM1 pKa = 7.18TMSDD5 pKa = 3.24EE6 pKa = 4.52SNNFDD11 pKa = 3.08EE12 pKa = 4.54TRR14 pKa = 11.84PIEE17 pKa = 4.76RR18 pKa = 11.84SPQPPAYY25 pKa = 9.87DD26 pKa = 3.83PYY28 pKa = 10.88GNPYY32 pKa = 9.01QDD34 pKa = 3.93PYY36 pKa = 11.46GGPFQQAAPHH46 pKa = 6.21PAPVRR51 pKa = 11.84AARR54 pKa = 11.84SGWSWRR60 pKa = 11.84SSLAGAVAGGVIAASVAVPVSWALASGGGQSTVADD95 pKa = 4.44APATAPQQLPEE106 pKa = 5.25PGDD109 pKa = 3.81SGSSGQAPDD118 pKa = 3.39TGQQGGTDD126 pKa = 3.6PFGYY130 pKa = 9.87GAPGGQLGGSTSNQTGDD147 pKa = 3.44QTDD150 pKa = 3.29ASADD154 pKa = 3.48QSQGVVLIDD163 pKa = 3.55TEE165 pKa = 4.65TTAGEE170 pKa = 3.92AAGTGLVLDD179 pKa = 3.99SSGIVLTNYY188 pKa = 9.94HH189 pKa = 5.37VVEE192 pKa = 4.87GSTSVKK198 pKa = 9.0VTIATTGATYY208 pKa = 9.98DD209 pKa = 3.8AKK211 pKa = 11.29VLGHH215 pKa = 6.24DD216 pKa = 3.69QEE218 pKa = 6.52ADD220 pKa = 3.49VALLQLDD227 pKa = 4.42GASGLATVKK236 pKa = 10.62LDD238 pKa = 4.46DD239 pKa = 5.62DD240 pKa = 4.81GDD242 pKa = 4.03PAVSDD247 pKa = 3.55EE248 pKa = 4.17VTAVGNAQGQGYY260 pKa = 9.8LSASTGSVVDD270 pKa = 4.76LDD272 pKa = 4.18QSIDD276 pKa = 3.67TQSEE280 pKa = 4.53GTVEE284 pKa = 4.45GEE286 pKa = 4.09HH287 pKa = 5.69LTGLIEE293 pKa = 4.0TNAYY297 pKa = 9.36VVGGYY302 pKa = 9.85SGGALLDD309 pKa = 3.82SEE311 pKa = 5.11GEE313 pKa = 4.39VVGITTAASSGGVAQSYY330 pKa = 9.54AVPIEE335 pKa = 4.56DD336 pKa = 4.18ALQVVQKK343 pKa = 10.52IEE345 pKa = 4.76DD346 pKa = 3.63GSEE349 pKa = 3.85TDD351 pKa = 3.73GVQVGASPYY360 pKa = 10.18LGIAISQDD368 pKa = 3.0ATGTSGGVEE377 pKa = 4.03VGQVQSGGAAAQAGIEE393 pKa = 4.09AGATITGIDD402 pKa = 3.42GTAITSYY409 pKa = 11.38DD410 pKa = 3.63VLKK413 pKa = 10.16STLATYY419 pKa = 10.49QPGDD423 pKa = 3.64SVTLRR428 pKa = 11.84WADD431 pKa = 3.4TSGATHH437 pKa = 6.79SAQVTLGEE445 pKa = 4.44SPVNN449 pKa = 3.57
MM1 pKa = 7.18TMSDD5 pKa = 3.24EE6 pKa = 4.52SNNFDD11 pKa = 3.08EE12 pKa = 4.54TRR14 pKa = 11.84PIEE17 pKa = 4.76RR18 pKa = 11.84SPQPPAYY25 pKa = 9.87DD26 pKa = 3.83PYY28 pKa = 10.88GNPYY32 pKa = 9.01QDD34 pKa = 3.93PYY36 pKa = 11.46GGPFQQAAPHH46 pKa = 6.21PAPVRR51 pKa = 11.84AARR54 pKa = 11.84SGWSWRR60 pKa = 11.84SSLAGAVAGGVIAASVAVPVSWALASGGGQSTVADD95 pKa = 4.44APATAPQQLPEE106 pKa = 5.25PGDD109 pKa = 3.81SGSSGQAPDD118 pKa = 3.39TGQQGGTDD126 pKa = 3.6PFGYY130 pKa = 9.87GAPGGQLGGSTSNQTGDD147 pKa = 3.44QTDD150 pKa = 3.29ASADD154 pKa = 3.48QSQGVVLIDD163 pKa = 3.55TEE165 pKa = 4.65TTAGEE170 pKa = 3.92AAGTGLVLDD179 pKa = 3.99SSGIVLTNYY188 pKa = 9.94HH189 pKa = 5.37VVEE192 pKa = 4.87GSTSVKK198 pKa = 9.0VTIATTGATYY208 pKa = 9.98DD209 pKa = 3.8AKK211 pKa = 11.29VLGHH215 pKa = 6.24DD216 pKa = 3.69QEE218 pKa = 6.52ADD220 pKa = 3.49VALLQLDD227 pKa = 4.42GASGLATVKK236 pKa = 10.62LDD238 pKa = 4.46DD239 pKa = 5.62DD240 pKa = 4.81GDD242 pKa = 4.03PAVSDD247 pKa = 3.55EE248 pKa = 4.17VTAVGNAQGQGYY260 pKa = 9.8LSASTGSVVDD270 pKa = 4.76LDD272 pKa = 4.18QSIDD276 pKa = 3.67TQSEE280 pKa = 4.53GTVEE284 pKa = 4.45GEE286 pKa = 4.09HH287 pKa = 5.69LTGLIEE293 pKa = 4.0TNAYY297 pKa = 9.36VVGGYY302 pKa = 9.85SGGALLDD309 pKa = 3.82SEE311 pKa = 5.11GEE313 pKa = 4.39VVGITTAASSGGVAQSYY330 pKa = 9.54AVPIEE335 pKa = 4.56DD336 pKa = 4.18ALQVVQKK343 pKa = 10.52IEE345 pKa = 4.76DD346 pKa = 3.63GSEE349 pKa = 3.85TDD351 pKa = 3.73GVQVGASPYY360 pKa = 10.18LGIAISQDD368 pKa = 3.0ATGTSGGVEE377 pKa = 4.03VGQVQSGGAAAQAGIEE393 pKa = 4.09AGATITGIDD402 pKa = 3.42GTAITSYY409 pKa = 11.38DD410 pKa = 3.63VLKK413 pKa = 10.16STLATYY419 pKa = 10.49QPGDD423 pKa = 3.64SVTLRR428 pKa = 11.84WADD431 pKa = 3.4TSGATHH437 pKa = 6.79SAQVTLGEE445 pKa = 4.44SPVNN449 pKa = 3.57
Molecular weight: 44.55 kDa
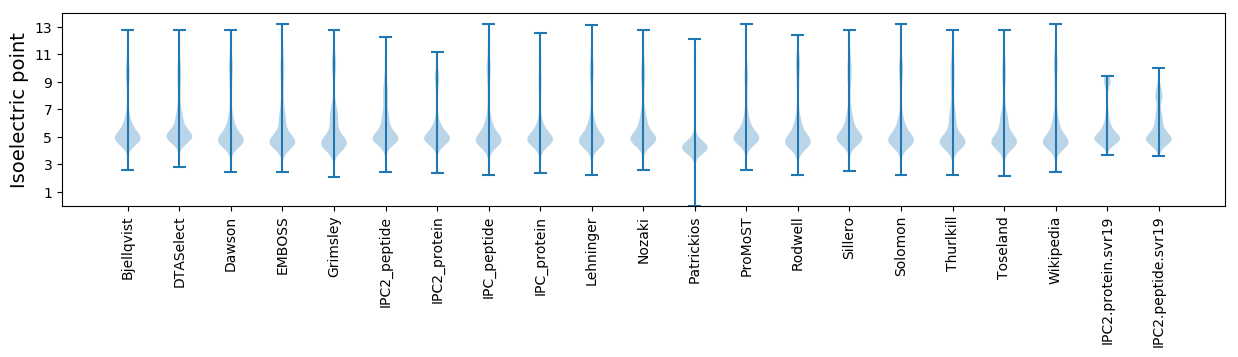
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7BPE0|A0A0Q7BPE0_9ACTN Uncharacterized protein OS=Nocardioides sp. Root1257 OX=1736439 GN=ASC77_12140 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSSRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.24GRR40 pKa = 11.84KK41 pKa = 8.84SLAVV45 pKa = 3.3
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSSRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.24GRR40 pKa = 11.84KK41 pKa = 8.84SLAVV45 pKa = 3.3
Molecular weight: 5.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1661918 |
35 |
2517 |
328.2 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.969 ± 0.05 | 0.704 ± 0.008 |
6.68 ± 0.028 | 5.517 ± 0.035 |
2.811 ± 0.019 | 9.212 ± 0.033 |
2.158 ± 0.018 | 3.566 ± 0.028 |
1.97 ± 0.029 | 10.155 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.842 ± 0.015 | 1.785 ± 0.022 |
5.549 ± 0.03 | 2.79 ± 0.018 |
7.313 ± 0.048 | 5.409 ± 0.028 |
6.391 ± 0.043 | 9.552 ± 0.033 |
1.566 ± 0.014 | 2.061 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
