
Robertkochia marina
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Robertkochia
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
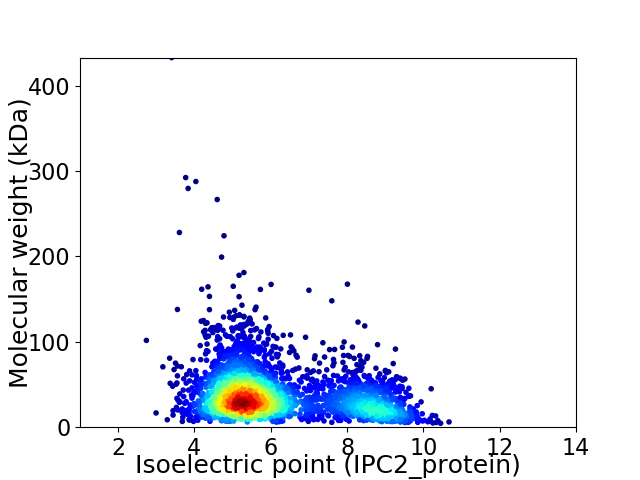
Virtual 2D-PAGE plot for 2974 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S3M1S0|A0A4S3M1S0_9FLAO Transporter OS=Robertkochia marina OX=1227945 GN=E7Z59_01065 PE=4 SV=1
MM1 pKa = 7.66AKK3 pKa = 10.39LLPRR7 pKa = 11.84FSWRR11 pKa = 11.84KK12 pKa = 8.98SFSSYY17 pKa = 10.76SLHH20 pKa = 7.29AILFSVFLLISTTALRR36 pKa = 11.84SQSVSFGQTGLQGEE50 pKa = 4.76NVTNPTSLQFGPNGKK65 pKa = 10.13LYY67 pKa = 10.61VAQQDD72 pKa = 3.67GTIWEE77 pKa = 4.28MVIEE81 pKa = 4.42RR82 pKa = 11.84DD83 pKa = 3.23EE84 pKa = 4.29AAPGSGSYY92 pKa = 8.92TVTNQNAITIIKK104 pKa = 10.5NEE106 pKa = 4.29TPNHH110 pKa = 6.29NDD112 pKa = 3.41DD113 pKa = 3.67GTINSDD119 pKa = 3.16NVRR122 pKa = 11.84QITGIMVTGTAEE134 pKa = 3.96NPILYY139 pKa = 8.11VTSSDD144 pKa = 3.46YY145 pKa = 11.0RR146 pKa = 11.84IGGGPDD152 pKa = 3.08LGDD155 pKa = 5.06DD156 pKa = 4.12RR157 pKa = 11.84NLDD160 pKa = 3.76TNSGVLSRR168 pKa = 11.84LTWDD172 pKa = 3.5GSNWDD177 pKa = 3.52KK178 pKa = 11.51VDD180 pKa = 3.97LVRR183 pKa = 11.84GLPRR187 pKa = 11.84CEE189 pKa = 4.49EE190 pKa = 3.9NHH192 pKa = 5.34STNGISEE199 pKa = 4.15FEE201 pKa = 4.21RR202 pKa = 11.84NGRR205 pKa = 11.84NYY207 pKa = 11.1LLIQQGGNTNMGAPSNNFAGAPEE230 pKa = 4.3TFLSGALLIIDD241 pKa = 4.41LTQLEE246 pKa = 4.53EE247 pKa = 4.21MEE249 pKa = 4.42TAAGGPYY256 pKa = 10.15LDD258 pKa = 4.7PRR260 pKa = 11.84TGNTPFVYY268 pKa = 10.41DD269 pKa = 5.2LPTLNDD275 pKa = 3.99PEE277 pKa = 5.36RR278 pKa = 11.84MDD280 pKa = 3.51ITNSHH285 pKa = 7.06PSFPYY290 pKa = 10.08SATHH294 pKa = 6.4PLYY297 pKa = 10.89NATIDD302 pKa = 3.87LGDD305 pKa = 3.84PFGGNNGLNQAFTEE319 pKa = 4.29VGGPVQIFSPGYY331 pKa = 7.76RR332 pKa = 11.84NAYY335 pKa = 9.81DD336 pKa = 3.52VVITQGGLIYY346 pKa = 10.62TSDD349 pKa = 3.72NGPNATWGGPPVIYY363 pKa = 10.57DD364 pKa = 3.32NSNTVKK370 pKa = 10.59GDD372 pKa = 3.62NSNTTYY378 pKa = 10.86EE379 pKa = 4.34PALGDD384 pKa = 3.54YY385 pKa = 7.65VTNQFNLTSGITHH398 pKa = 7.48GDD400 pKa = 3.16QLHH403 pKa = 6.41YY404 pKa = 10.45IGTINDD410 pKa = 3.87PNGTYY415 pKa = 10.46YY416 pKa = 10.67GGHH419 pKa = 7.01PAPVRR424 pKa = 11.84AFPSRR429 pKa = 11.84AGIIKK434 pKa = 10.01YY435 pKa = 9.87VYY437 pKa = 10.17QGGNWINDD445 pKa = 3.7GAYY448 pKa = 10.46SLSDD452 pKa = 3.96LLDD455 pKa = 3.75GTSGYY460 pKa = 10.2FKK462 pKa = 10.95SSFSLADD469 pKa = 3.74FPDD472 pKa = 4.29DD473 pKa = 4.24PRR475 pKa = 11.84QGDD478 pKa = 3.67YY479 pKa = 7.57TTNEE483 pKa = 3.57ISNPDD488 pKa = 4.06LNILDD493 pKa = 3.82IVGSSTNGICEE504 pKa = 4.17FTASNFNGAMQGDD517 pKa = 5.49LLTASWSGKK526 pKa = 7.34INRR529 pKa = 11.84YY530 pKa = 9.32KK531 pKa = 10.71LGAGGTTLVSKK542 pKa = 11.0NNSFLSGFGSQPLDD556 pKa = 5.14LIAQGDD562 pKa = 3.81NAIFPGTIWAVTFGANNITIFEE584 pKa = 4.3PADD587 pKa = 3.61FVSCLLPGDD596 pKa = 4.42PDD598 pKa = 3.66YY599 pKa = 11.48DD600 pKa = 4.19ASLDD604 pKa = 3.84YY605 pKa = 11.64DD606 pKa = 3.51NDD608 pKa = 4.17GYY610 pKa = 9.84TNEE613 pKa = 5.04DD614 pKa = 3.3EE615 pKa = 4.61VLNGTDD621 pKa = 3.34LCSGGSRR628 pKa = 11.84PNDD631 pKa = 3.25FDD633 pKa = 4.28EE634 pKa = 6.72DD635 pKa = 4.39GVSDD639 pKa = 5.9LLDD642 pKa = 5.16DD643 pKa = 6.34DD644 pKa = 5.78DD645 pKa = 6.86DD646 pKa = 5.8NDD648 pKa = 5.6GIPDD652 pKa = 3.56IQDD655 pKa = 2.92AFAIDD660 pKa = 3.7AANGLTTNLPVQFPFWNNDD679 pKa = 2.97PGTGFFGLGFTGLMLDD695 pKa = 4.23PDD697 pKa = 4.8FSTDD701 pKa = 3.34YY702 pKa = 10.84LNQYY706 pKa = 9.2VEE708 pKa = 4.68EE709 pKa = 4.16NLSFGGAAGKK719 pKa = 8.91ATVDD723 pKa = 3.64VVSPGSALTSQNDD736 pKa = 2.97QDD738 pKa = 3.88YY739 pKa = 10.39GFQFGVNVDD748 pKa = 3.81EE749 pKa = 4.74NSNPFTIQVGIDD761 pKa = 3.25TPFNGTLPEE770 pKa = 4.39SGQEE774 pKa = 3.75YY775 pKa = 10.64GVFIGNGDD783 pKa = 3.27QDD785 pKa = 3.91NYY787 pKa = 10.79LKK789 pKa = 10.96LVISEE794 pKa = 4.17GQNANDD800 pKa = 3.64GLAGFNLLLEE810 pKa = 5.0DD811 pKa = 4.18NGTIINTQKK820 pKa = 10.55FDD822 pKa = 3.32VAGILEE828 pKa = 4.53ASDD831 pKa = 3.73MILYY835 pKa = 10.54LGVNPSLGEE844 pKa = 3.79VQAYY848 pKa = 9.26YY849 pKa = 10.7SIDD852 pKa = 3.32GGLSIEE858 pKa = 4.38SLGTAVTLPGTFLNTADD875 pKa = 3.99AKK877 pKa = 10.71GLAVGLISTSGQAPSFTATWDD898 pKa = 4.34FINITEE904 pKa = 4.41DD905 pKa = 3.36QPGVLQANTTSLDD918 pKa = 3.35FGTLQINSAQNNLSLDD934 pKa = 3.63LTNLGGATAPKK945 pKa = 10.1IAVTEE950 pKa = 4.41LNFSGTDD957 pKa = 3.12ALLFDD962 pKa = 4.93TNASLPLEE970 pKa = 4.44IGPGEE975 pKa = 4.29TKK977 pKa = 10.34IIPVNILPDD986 pKa = 3.69SEE988 pKa = 4.32PRR990 pKa = 11.84TVSASLEE997 pKa = 4.38VVHH1000 pKa = 6.87SGTNSPLDD1008 pKa = 3.66FPLTAEE1014 pKa = 4.11FVEE1017 pKa = 4.48VLNTTVIRR1025 pKa = 11.84VNTGGGQEE1033 pKa = 4.04NYY1035 pKa = 9.19EE1036 pKa = 4.38SKK1038 pKa = 11.0VFEE1041 pKa = 4.53ADD1043 pKa = 3.23QFFTGGKK1050 pKa = 10.0VYY1052 pKa = 10.66TNNQADD1058 pKa = 3.91VPTLFKK1064 pKa = 10.55SEE1066 pKa = 4.34RR1067 pKa = 11.84SASPPQFAYY1076 pKa = 10.68NFPVPDD1082 pKa = 3.51GDD1084 pKa = 4.11YY1085 pKa = 10.91QVNLYY1090 pKa = 9.79FAEE1093 pKa = 4.64IYY1095 pKa = 9.58WGATGGGTGGIGSRR1109 pKa = 11.84IFDD1112 pKa = 3.4VTIEE1116 pKa = 4.38DD1117 pKa = 4.09NLVLDD1122 pKa = 5.05DD1123 pKa = 5.0LDD1125 pKa = 3.69IFAEE1129 pKa = 4.33VGAEE1133 pKa = 4.08SVLVKK1138 pKa = 10.04PFNVVVADD1146 pKa = 4.0GQLNIAFNAASAVGGNDD1163 pKa = 3.08QPKK1166 pKa = 9.89ISAIEE1171 pKa = 3.81ILSTQNKK1178 pKa = 8.6IPNPQISSSVTSGPAPLFVDD1198 pKa = 4.87FNGSGSSDD1206 pKa = 3.72DD1207 pKa = 4.77DD1208 pKa = 5.03GIIAYY1213 pKa = 9.59SWDD1216 pKa = 3.67FGDD1219 pKa = 4.73GSVSEE1224 pKa = 4.84QINPSNIFTTPGLYY1238 pKa = 9.08TVSLTVTDD1246 pKa = 3.05TRR1248 pKa = 11.84GAVNTTSVEE1257 pKa = 4.01IEE1259 pKa = 4.1VTDD1262 pKa = 3.99GNNNDD1267 pKa = 2.82FSLRR1271 pKa = 11.84VNAGGSSVNYY1281 pKa = 10.02AGKK1284 pKa = 8.74TYY1286 pKa = 10.99LNDD1289 pKa = 3.27QYY1291 pKa = 10.38FTNGTSFQNTNATVPDD1307 pKa = 3.98LFITEE1312 pKa = 4.27RR1313 pKa = 11.84TSPLDD1318 pKa = 3.29FQYY1321 pKa = 11.27NFPVANGDD1329 pKa = 3.99YY1330 pKa = 10.69LVSLYY1335 pKa = 10.34FAEE1338 pKa = 5.07IYY1340 pKa = 9.59WGAPGGKK1347 pKa = 9.02EE1348 pKa = 3.8GGVGSRR1354 pKa = 11.84VFDD1357 pKa = 3.74VEE1359 pKa = 4.33MEE1361 pKa = 4.2GATILDD1367 pKa = 4.14DD1368 pKa = 3.87FDD1370 pKa = 4.86IYY1372 pKa = 11.53AEE1374 pKa = 4.47AGAEE1378 pKa = 4.18VPINRR1383 pKa = 11.84SFVATVNDD1391 pKa = 5.39GILDD1395 pKa = 3.94LVFTSDD1401 pKa = 3.44PSKK1404 pKa = 11.35GGVDD1408 pKa = 3.57QPKK1411 pKa = 9.91VSAIEE1416 pKa = 4.06IIGVDD1421 pKa = 3.31GGNFPVLTLLDD1432 pKa = 3.84IPDD1435 pKa = 3.65RR1436 pKa = 11.84TSVVDD1441 pKa = 3.5EE1442 pKa = 4.82VIDD1445 pKa = 4.04FGVSASGGDD1454 pKa = 3.37PSEE1457 pKa = 4.76NITYY1461 pKa = 10.29AISGQPNGVDD1471 pKa = 3.06IEE1473 pKa = 4.36PTNGQIFGVIEE1484 pKa = 4.1SSALQGGPQGNGVHH1498 pKa = 6.01NVTVTVSQPSSNTVEE1513 pKa = 4.76KK1514 pKa = 10.16QFEE1517 pKa = 4.81WIVTPANSSWTLKK1530 pKa = 10.22PQVEE1534 pKa = 5.04NYY1536 pKa = 7.12TARR1539 pKa = 11.84HH1540 pKa = 5.14EE1541 pKa = 4.55CGFVQAGTKK1550 pKa = 10.14FYY1552 pKa = 11.04LLGGRR1557 pKa = 11.84EE1558 pKa = 4.24SATTMEE1564 pKa = 4.41VYY1566 pKa = 10.33DD1567 pKa = 4.1YY1568 pKa = 9.51QTNIWSALQTVAPKK1582 pKa = 10.48DD1583 pKa = 3.77FNHH1586 pKa = 6.26IQPVLYY1592 pKa = 10.46DD1593 pKa = 3.38GLIWIVGGFQTNNYY1607 pKa = 8.1PNEE1610 pKa = 3.94VSLDD1614 pKa = 3.73HH1615 pKa = 6.75VWIFNPATEE1624 pKa = 3.85SWYY1627 pKa = 10.21QGPAIPEE1634 pKa = 3.86GRR1636 pKa = 11.84KK1637 pKa = 9.49RR1638 pKa = 11.84GSAGTVVYY1646 pKa = 10.67NNKK1649 pKa = 9.76IYY1651 pKa = 10.85LIGGNTQGHH1660 pKa = 6.99DD1661 pKa = 3.54GGYY1664 pKa = 10.05VPYY1667 pKa = 10.51FDD1669 pKa = 5.22VFDD1672 pKa = 4.58PATGKK1677 pKa = 7.22WTSLPDD1683 pKa = 3.19APRR1686 pKa = 11.84ARR1688 pKa = 11.84DD1689 pKa = 3.32HH1690 pKa = 6.14FQAVVINGKK1699 pKa = 10.06LYY1701 pKa = 10.49AAAGRR1706 pKa = 11.84LTGGEE1711 pKa = 3.99GGVFGPAIPEE1721 pKa = 3.59VDD1723 pKa = 3.56VYY1725 pKa = 11.78DD1726 pKa = 4.78FGSGTWTTLSQEE1738 pKa = 4.31KK1739 pKa = 10.46NIPTPRR1745 pKa = 11.84ASAASVAYY1753 pKa = 9.68NGKK1756 pKa = 8.91VYY1758 pKa = 11.19VMGGEE1763 pKa = 4.37AEE1765 pKa = 4.29SSAITQAFKK1774 pKa = 9.71ITEE1777 pKa = 4.14IYY1779 pKa = 10.63DD1780 pKa = 4.06PIIGSWSTGPEE1791 pKa = 4.0MNFGRR1796 pKa = 11.84HH1797 pKa = 4.06GTQAIVSGQGIHH1809 pKa = 5.72ITGGSPKK1816 pKa = 10.27RR1817 pKa = 11.84GGGNQKK1823 pKa = 10.31NMEE1826 pKa = 4.63YY1827 pKa = 10.99LNIDD1831 pKa = 3.77APQGNPSNASEE1842 pKa = 4.14LQVTEE1847 pKa = 4.05IVSFNPGEE1855 pKa = 4.24TKK1857 pKa = 10.08IVAFNITGGNVGVWLNSIQTIGANAGEE1884 pKa = 4.35FNVVSSDD1891 pKa = 3.21VSNRR1895 pKa = 11.84LLSSGNTYY1903 pKa = 10.07TITVKK1908 pKa = 9.39HH1909 pKa = 6.02TGSLDD1914 pKa = 3.6EE1915 pKa = 4.21VVVPLEE1921 pKa = 3.52ISFNDD1926 pKa = 4.01GEE1928 pKa = 4.39SAIINLVIGNPPIGDD1943 pKa = 3.52AVTSFSLVNADD1954 pKa = 3.78TNNDD1958 pKa = 3.37LFTINDD1964 pKa = 3.46GDD1966 pKa = 4.83LIDD1969 pKa = 3.87VNQIVGVGLNIRR1981 pKa = 11.84ANTNPSQVGSVVFSLQGPISVNKK2004 pKa = 9.53KK2005 pKa = 9.99EE2006 pKa = 4.35SVAPYY2011 pKa = 10.29AIFGDD2016 pKa = 3.56VSGNYY2021 pKa = 9.7NPGALPEE2028 pKa = 4.34GNYY2031 pKa = 9.99VLSATPYY2038 pKa = 10.36SEE2040 pKa = 4.71SKK2042 pKa = 11.42GNGTPGATLLINFSIGQPSDD2062 pKa = 5.18DD2063 pKa = 4.2IPPTALATADD2073 pKa = 3.3IVAGGVPLSVTFTGSGSSPGANAIAAYY2100 pKa = 9.46EE2101 pKa = 4.0WDD2103 pKa = 3.99FGDD2106 pKa = 4.46GNSSSEE2112 pKa = 3.96IDD2114 pKa = 3.3PTHH2117 pKa = 7.4IFEE2120 pKa = 4.21QSGVFTVTLLVTDD2133 pKa = 4.21TEE2135 pKa = 4.59GLSDD2139 pKa = 3.65TATLEE2144 pKa = 3.82ITVNDD2149 pKa = 4.21PVNNGPIAIATSDD2162 pKa = 3.3ITSGEE2167 pKa = 4.08VPLTVNFDD2175 pKa = 3.92GSGSTQGDD2183 pKa = 3.8NPIVSYY2189 pKa = 10.88AWDD2192 pKa = 4.03FGDD2195 pKa = 4.81GNTSGIAEE2203 pKa = 4.32PVHH2206 pKa = 6.9EE2207 pKa = 4.28YY2208 pKa = 8.34TAPGVYY2214 pKa = 8.91TAVLTVVDD2222 pKa = 4.27EE2223 pKa = 4.97LGLEE2227 pKa = 4.18STDD2230 pKa = 3.64EE2231 pKa = 4.21IEE2233 pKa = 3.96ITVNDD2238 pKa = 4.06IVPEE2242 pKa = 4.21GDD2244 pKa = 3.25AVISFTLINADD2255 pKa = 3.15TDD2257 pKa = 3.72ADD2259 pKa = 4.24LLILSEE2265 pKa = 4.59GIIVDD2270 pKa = 3.92ATAVNGTGLNIRR2282 pKa = 11.84ANTNPLQVGSVVLQLTGPVGNLRR2305 pKa = 11.84TEE2307 pKa = 4.23SVAPYY2312 pKa = 11.06ALFGDD2317 pKa = 3.64SGGNYY2322 pKa = 9.68FGQALPEE2329 pKa = 4.19GNYY2332 pKa = 9.95TITATPFSASGGGGTPGTPLTINFSIGDD2360 pKa = 3.77PVIVVGPTAIATSNLQSGEE2379 pKa = 4.14APLEE2383 pKa = 3.76ILFNGSGSLEE2393 pKa = 3.99GDD2395 pKa = 3.38NPIINYY2401 pKa = 9.69AWDD2404 pKa = 4.05FGDD2407 pKa = 4.56GGSSEE2412 pKa = 4.67QPDD2415 pKa = 3.44PTYY2418 pKa = 11.27NYY2420 pKa = 7.37NTPGNYY2426 pKa = 7.74TATLTVTDD2434 pKa = 4.44DD2435 pKa = 4.52QGLQDD2440 pKa = 4.32SDD2442 pKa = 3.57QLIISVSGPVITGPTAIAVSDD2463 pKa = 4.01VTSGEE2468 pKa = 4.06APFEE2472 pKa = 4.68INFNGSGSVQGDD2484 pKa = 3.68NPITTYY2490 pKa = 10.66SWDD2493 pKa = 3.7FGDD2496 pKa = 5.59GGTSDD2501 pKa = 5.12LADD2504 pKa = 3.42PTYY2507 pKa = 10.76TYY2509 pKa = 8.17LTPGSFLAVLTVTDD2523 pKa = 4.25DD2524 pKa = 3.75QGLQATDD2531 pKa = 4.2EE2532 pKa = 4.33INITVTPPAPEE2543 pKa = 4.64ADD2545 pKa = 3.27QVVSFTLINAATDD2558 pKa = 3.28NDD2560 pKa = 4.24LFDD2563 pKa = 3.52ITEE2566 pKa = 4.62GIIIDD2571 pKa = 4.35LNSIGTPSLNIRR2583 pKa = 11.84ANTSPAVVGSVIMQLSGPVSNSRR2606 pKa = 11.84TEE2608 pKa = 4.09SVAPYY2613 pKa = 10.91ALFGDD2618 pKa = 3.89SSGNYY2623 pKa = 9.11FNGSLTPGDD2632 pKa = 4.09YY2633 pKa = 11.01SLTATPYY2640 pKa = 8.36TAGGGGGTAGTPLVINFSVVNTQTKK2665 pKa = 10.09SGTIAPDD2672 pKa = 3.11SSLGEE2677 pKa = 3.95EE2678 pKa = 4.26TVLSEE2683 pKa = 5.19EE2684 pKa = 4.23VLLYY2688 pKa = 9.5PNPTNYY2694 pKa = 9.46TLNISGLSSTDD2705 pKa = 2.67ISQEE2709 pKa = 3.35ARR2711 pKa = 11.84FYY2713 pKa = 10.04MHH2715 pKa = 7.68DD2716 pKa = 3.21IKK2718 pKa = 11.24GALLKK2723 pKa = 10.03TFTPNGVEE2731 pKa = 4.18SADD2734 pKa = 3.94GYY2736 pKa = 11.45ALPVDD2741 pKa = 4.83DD2742 pKa = 5.14LQQGVYY2748 pKa = 10.51LLSLEE2753 pKa = 4.4SNGRR2757 pKa = 11.84VLLRR2761 pKa = 11.84KK2762 pKa = 9.59RR2763 pKa = 11.84FVVMKK2768 pKa = 9.76QQ2769 pKa = 2.79
MM1 pKa = 7.66AKK3 pKa = 10.39LLPRR7 pKa = 11.84FSWRR11 pKa = 11.84KK12 pKa = 8.98SFSSYY17 pKa = 10.76SLHH20 pKa = 7.29AILFSVFLLISTTALRR36 pKa = 11.84SQSVSFGQTGLQGEE50 pKa = 4.76NVTNPTSLQFGPNGKK65 pKa = 10.13LYY67 pKa = 10.61VAQQDD72 pKa = 3.67GTIWEE77 pKa = 4.28MVIEE81 pKa = 4.42RR82 pKa = 11.84DD83 pKa = 3.23EE84 pKa = 4.29AAPGSGSYY92 pKa = 8.92TVTNQNAITIIKK104 pKa = 10.5NEE106 pKa = 4.29TPNHH110 pKa = 6.29NDD112 pKa = 3.41DD113 pKa = 3.67GTINSDD119 pKa = 3.16NVRR122 pKa = 11.84QITGIMVTGTAEE134 pKa = 3.96NPILYY139 pKa = 8.11VTSSDD144 pKa = 3.46YY145 pKa = 11.0RR146 pKa = 11.84IGGGPDD152 pKa = 3.08LGDD155 pKa = 5.06DD156 pKa = 4.12RR157 pKa = 11.84NLDD160 pKa = 3.76TNSGVLSRR168 pKa = 11.84LTWDD172 pKa = 3.5GSNWDD177 pKa = 3.52KK178 pKa = 11.51VDD180 pKa = 3.97LVRR183 pKa = 11.84GLPRR187 pKa = 11.84CEE189 pKa = 4.49EE190 pKa = 3.9NHH192 pKa = 5.34STNGISEE199 pKa = 4.15FEE201 pKa = 4.21RR202 pKa = 11.84NGRR205 pKa = 11.84NYY207 pKa = 11.1LLIQQGGNTNMGAPSNNFAGAPEE230 pKa = 4.3TFLSGALLIIDD241 pKa = 4.41LTQLEE246 pKa = 4.53EE247 pKa = 4.21MEE249 pKa = 4.42TAAGGPYY256 pKa = 10.15LDD258 pKa = 4.7PRR260 pKa = 11.84TGNTPFVYY268 pKa = 10.41DD269 pKa = 5.2LPTLNDD275 pKa = 3.99PEE277 pKa = 5.36RR278 pKa = 11.84MDD280 pKa = 3.51ITNSHH285 pKa = 7.06PSFPYY290 pKa = 10.08SATHH294 pKa = 6.4PLYY297 pKa = 10.89NATIDD302 pKa = 3.87LGDD305 pKa = 3.84PFGGNNGLNQAFTEE319 pKa = 4.29VGGPVQIFSPGYY331 pKa = 7.76RR332 pKa = 11.84NAYY335 pKa = 9.81DD336 pKa = 3.52VVITQGGLIYY346 pKa = 10.62TSDD349 pKa = 3.72NGPNATWGGPPVIYY363 pKa = 10.57DD364 pKa = 3.32NSNTVKK370 pKa = 10.59GDD372 pKa = 3.62NSNTTYY378 pKa = 10.86EE379 pKa = 4.34PALGDD384 pKa = 3.54YY385 pKa = 7.65VTNQFNLTSGITHH398 pKa = 7.48GDD400 pKa = 3.16QLHH403 pKa = 6.41YY404 pKa = 10.45IGTINDD410 pKa = 3.87PNGTYY415 pKa = 10.46YY416 pKa = 10.67GGHH419 pKa = 7.01PAPVRR424 pKa = 11.84AFPSRR429 pKa = 11.84AGIIKK434 pKa = 10.01YY435 pKa = 9.87VYY437 pKa = 10.17QGGNWINDD445 pKa = 3.7GAYY448 pKa = 10.46SLSDD452 pKa = 3.96LLDD455 pKa = 3.75GTSGYY460 pKa = 10.2FKK462 pKa = 10.95SSFSLADD469 pKa = 3.74FPDD472 pKa = 4.29DD473 pKa = 4.24PRR475 pKa = 11.84QGDD478 pKa = 3.67YY479 pKa = 7.57TTNEE483 pKa = 3.57ISNPDD488 pKa = 4.06LNILDD493 pKa = 3.82IVGSSTNGICEE504 pKa = 4.17FTASNFNGAMQGDD517 pKa = 5.49LLTASWSGKK526 pKa = 7.34INRR529 pKa = 11.84YY530 pKa = 9.32KK531 pKa = 10.71LGAGGTTLVSKK542 pKa = 11.0NNSFLSGFGSQPLDD556 pKa = 5.14LIAQGDD562 pKa = 3.81NAIFPGTIWAVTFGANNITIFEE584 pKa = 4.3PADD587 pKa = 3.61FVSCLLPGDD596 pKa = 4.42PDD598 pKa = 3.66YY599 pKa = 11.48DD600 pKa = 4.19ASLDD604 pKa = 3.84YY605 pKa = 11.64DD606 pKa = 3.51NDD608 pKa = 4.17GYY610 pKa = 9.84TNEE613 pKa = 5.04DD614 pKa = 3.3EE615 pKa = 4.61VLNGTDD621 pKa = 3.34LCSGGSRR628 pKa = 11.84PNDD631 pKa = 3.25FDD633 pKa = 4.28EE634 pKa = 6.72DD635 pKa = 4.39GVSDD639 pKa = 5.9LLDD642 pKa = 5.16DD643 pKa = 6.34DD644 pKa = 5.78DD645 pKa = 6.86DD646 pKa = 5.8NDD648 pKa = 5.6GIPDD652 pKa = 3.56IQDD655 pKa = 2.92AFAIDD660 pKa = 3.7AANGLTTNLPVQFPFWNNDD679 pKa = 2.97PGTGFFGLGFTGLMLDD695 pKa = 4.23PDD697 pKa = 4.8FSTDD701 pKa = 3.34YY702 pKa = 10.84LNQYY706 pKa = 9.2VEE708 pKa = 4.68EE709 pKa = 4.16NLSFGGAAGKK719 pKa = 8.91ATVDD723 pKa = 3.64VVSPGSALTSQNDD736 pKa = 2.97QDD738 pKa = 3.88YY739 pKa = 10.39GFQFGVNVDD748 pKa = 3.81EE749 pKa = 4.74NSNPFTIQVGIDD761 pKa = 3.25TPFNGTLPEE770 pKa = 4.39SGQEE774 pKa = 3.75YY775 pKa = 10.64GVFIGNGDD783 pKa = 3.27QDD785 pKa = 3.91NYY787 pKa = 10.79LKK789 pKa = 10.96LVISEE794 pKa = 4.17GQNANDD800 pKa = 3.64GLAGFNLLLEE810 pKa = 5.0DD811 pKa = 4.18NGTIINTQKK820 pKa = 10.55FDD822 pKa = 3.32VAGILEE828 pKa = 4.53ASDD831 pKa = 3.73MILYY835 pKa = 10.54LGVNPSLGEE844 pKa = 3.79VQAYY848 pKa = 9.26YY849 pKa = 10.7SIDD852 pKa = 3.32GGLSIEE858 pKa = 4.38SLGTAVTLPGTFLNTADD875 pKa = 3.99AKK877 pKa = 10.71GLAVGLISTSGQAPSFTATWDD898 pKa = 4.34FINITEE904 pKa = 4.41DD905 pKa = 3.36QPGVLQANTTSLDD918 pKa = 3.35FGTLQINSAQNNLSLDD934 pKa = 3.63LTNLGGATAPKK945 pKa = 10.1IAVTEE950 pKa = 4.41LNFSGTDD957 pKa = 3.12ALLFDD962 pKa = 4.93TNASLPLEE970 pKa = 4.44IGPGEE975 pKa = 4.29TKK977 pKa = 10.34IIPVNILPDD986 pKa = 3.69SEE988 pKa = 4.32PRR990 pKa = 11.84TVSASLEE997 pKa = 4.38VVHH1000 pKa = 6.87SGTNSPLDD1008 pKa = 3.66FPLTAEE1014 pKa = 4.11FVEE1017 pKa = 4.48VLNTTVIRR1025 pKa = 11.84VNTGGGQEE1033 pKa = 4.04NYY1035 pKa = 9.19EE1036 pKa = 4.38SKK1038 pKa = 11.0VFEE1041 pKa = 4.53ADD1043 pKa = 3.23QFFTGGKK1050 pKa = 10.0VYY1052 pKa = 10.66TNNQADD1058 pKa = 3.91VPTLFKK1064 pKa = 10.55SEE1066 pKa = 4.34RR1067 pKa = 11.84SASPPQFAYY1076 pKa = 10.68NFPVPDD1082 pKa = 3.51GDD1084 pKa = 4.11YY1085 pKa = 10.91QVNLYY1090 pKa = 9.79FAEE1093 pKa = 4.64IYY1095 pKa = 9.58WGATGGGTGGIGSRR1109 pKa = 11.84IFDD1112 pKa = 3.4VTIEE1116 pKa = 4.38DD1117 pKa = 4.09NLVLDD1122 pKa = 5.05DD1123 pKa = 5.0LDD1125 pKa = 3.69IFAEE1129 pKa = 4.33VGAEE1133 pKa = 4.08SVLVKK1138 pKa = 10.04PFNVVVADD1146 pKa = 4.0GQLNIAFNAASAVGGNDD1163 pKa = 3.08QPKK1166 pKa = 9.89ISAIEE1171 pKa = 3.81ILSTQNKK1178 pKa = 8.6IPNPQISSSVTSGPAPLFVDD1198 pKa = 4.87FNGSGSSDD1206 pKa = 3.72DD1207 pKa = 4.77DD1208 pKa = 5.03GIIAYY1213 pKa = 9.59SWDD1216 pKa = 3.67FGDD1219 pKa = 4.73GSVSEE1224 pKa = 4.84QINPSNIFTTPGLYY1238 pKa = 9.08TVSLTVTDD1246 pKa = 3.05TRR1248 pKa = 11.84GAVNTTSVEE1257 pKa = 4.01IEE1259 pKa = 4.1VTDD1262 pKa = 3.99GNNNDD1267 pKa = 2.82FSLRR1271 pKa = 11.84VNAGGSSVNYY1281 pKa = 10.02AGKK1284 pKa = 8.74TYY1286 pKa = 10.99LNDD1289 pKa = 3.27QYY1291 pKa = 10.38FTNGTSFQNTNATVPDD1307 pKa = 3.98LFITEE1312 pKa = 4.27RR1313 pKa = 11.84TSPLDD1318 pKa = 3.29FQYY1321 pKa = 11.27NFPVANGDD1329 pKa = 3.99YY1330 pKa = 10.69LVSLYY1335 pKa = 10.34FAEE1338 pKa = 5.07IYY1340 pKa = 9.59WGAPGGKK1347 pKa = 9.02EE1348 pKa = 3.8GGVGSRR1354 pKa = 11.84VFDD1357 pKa = 3.74VEE1359 pKa = 4.33MEE1361 pKa = 4.2GATILDD1367 pKa = 4.14DD1368 pKa = 3.87FDD1370 pKa = 4.86IYY1372 pKa = 11.53AEE1374 pKa = 4.47AGAEE1378 pKa = 4.18VPINRR1383 pKa = 11.84SFVATVNDD1391 pKa = 5.39GILDD1395 pKa = 3.94LVFTSDD1401 pKa = 3.44PSKK1404 pKa = 11.35GGVDD1408 pKa = 3.57QPKK1411 pKa = 9.91VSAIEE1416 pKa = 4.06IIGVDD1421 pKa = 3.31GGNFPVLTLLDD1432 pKa = 3.84IPDD1435 pKa = 3.65RR1436 pKa = 11.84TSVVDD1441 pKa = 3.5EE1442 pKa = 4.82VIDD1445 pKa = 4.04FGVSASGGDD1454 pKa = 3.37PSEE1457 pKa = 4.76NITYY1461 pKa = 10.29AISGQPNGVDD1471 pKa = 3.06IEE1473 pKa = 4.36PTNGQIFGVIEE1484 pKa = 4.1SSALQGGPQGNGVHH1498 pKa = 6.01NVTVTVSQPSSNTVEE1513 pKa = 4.76KK1514 pKa = 10.16QFEE1517 pKa = 4.81WIVTPANSSWTLKK1530 pKa = 10.22PQVEE1534 pKa = 5.04NYY1536 pKa = 7.12TARR1539 pKa = 11.84HH1540 pKa = 5.14EE1541 pKa = 4.55CGFVQAGTKK1550 pKa = 10.14FYY1552 pKa = 11.04LLGGRR1557 pKa = 11.84EE1558 pKa = 4.24SATTMEE1564 pKa = 4.41VYY1566 pKa = 10.33DD1567 pKa = 4.1YY1568 pKa = 9.51QTNIWSALQTVAPKK1582 pKa = 10.48DD1583 pKa = 3.77FNHH1586 pKa = 6.26IQPVLYY1592 pKa = 10.46DD1593 pKa = 3.38GLIWIVGGFQTNNYY1607 pKa = 8.1PNEE1610 pKa = 3.94VSLDD1614 pKa = 3.73HH1615 pKa = 6.75VWIFNPATEE1624 pKa = 3.85SWYY1627 pKa = 10.21QGPAIPEE1634 pKa = 3.86GRR1636 pKa = 11.84KK1637 pKa = 9.49RR1638 pKa = 11.84GSAGTVVYY1646 pKa = 10.67NNKK1649 pKa = 9.76IYY1651 pKa = 10.85LIGGNTQGHH1660 pKa = 6.99DD1661 pKa = 3.54GGYY1664 pKa = 10.05VPYY1667 pKa = 10.51FDD1669 pKa = 5.22VFDD1672 pKa = 4.58PATGKK1677 pKa = 7.22WTSLPDD1683 pKa = 3.19APRR1686 pKa = 11.84ARR1688 pKa = 11.84DD1689 pKa = 3.32HH1690 pKa = 6.14FQAVVINGKK1699 pKa = 10.06LYY1701 pKa = 10.49AAAGRR1706 pKa = 11.84LTGGEE1711 pKa = 3.99GGVFGPAIPEE1721 pKa = 3.59VDD1723 pKa = 3.56VYY1725 pKa = 11.78DD1726 pKa = 4.78FGSGTWTTLSQEE1738 pKa = 4.31KK1739 pKa = 10.46NIPTPRR1745 pKa = 11.84ASAASVAYY1753 pKa = 9.68NGKK1756 pKa = 8.91VYY1758 pKa = 11.19VMGGEE1763 pKa = 4.37AEE1765 pKa = 4.29SSAITQAFKK1774 pKa = 9.71ITEE1777 pKa = 4.14IYY1779 pKa = 10.63DD1780 pKa = 4.06PIIGSWSTGPEE1791 pKa = 4.0MNFGRR1796 pKa = 11.84HH1797 pKa = 4.06GTQAIVSGQGIHH1809 pKa = 5.72ITGGSPKK1816 pKa = 10.27RR1817 pKa = 11.84GGGNQKK1823 pKa = 10.31NMEE1826 pKa = 4.63YY1827 pKa = 10.99LNIDD1831 pKa = 3.77APQGNPSNASEE1842 pKa = 4.14LQVTEE1847 pKa = 4.05IVSFNPGEE1855 pKa = 4.24TKK1857 pKa = 10.08IVAFNITGGNVGVWLNSIQTIGANAGEE1884 pKa = 4.35FNVVSSDD1891 pKa = 3.21VSNRR1895 pKa = 11.84LLSSGNTYY1903 pKa = 10.07TITVKK1908 pKa = 9.39HH1909 pKa = 6.02TGSLDD1914 pKa = 3.6EE1915 pKa = 4.21VVVPLEE1921 pKa = 3.52ISFNDD1926 pKa = 4.01GEE1928 pKa = 4.39SAIINLVIGNPPIGDD1943 pKa = 3.52AVTSFSLVNADD1954 pKa = 3.78TNNDD1958 pKa = 3.37LFTINDD1964 pKa = 3.46GDD1966 pKa = 4.83LIDD1969 pKa = 3.87VNQIVGVGLNIRR1981 pKa = 11.84ANTNPSQVGSVVFSLQGPISVNKK2004 pKa = 9.53KK2005 pKa = 9.99EE2006 pKa = 4.35SVAPYY2011 pKa = 10.29AIFGDD2016 pKa = 3.56VSGNYY2021 pKa = 9.7NPGALPEE2028 pKa = 4.34GNYY2031 pKa = 9.99VLSATPYY2038 pKa = 10.36SEE2040 pKa = 4.71SKK2042 pKa = 11.42GNGTPGATLLINFSIGQPSDD2062 pKa = 5.18DD2063 pKa = 4.2IPPTALATADD2073 pKa = 3.3IVAGGVPLSVTFTGSGSSPGANAIAAYY2100 pKa = 9.46EE2101 pKa = 4.0WDD2103 pKa = 3.99FGDD2106 pKa = 4.46GNSSSEE2112 pKa = 3.96IDD2114 pKa = 3.3PTHH2117 pKa = 7.4IFEE2120 pKa = 4.21QSGVFTVTLLVTDD2133 pKa = 4.21TEE2135 pKa = 4.59GLSDD2139 pKa = 3.65TATLEE2144 pKa = 3.82ITVNDD2149 pKa = 4.21PVNNGPIAIATSDD2162 pKa = 3.3ITSGEE2167 pKa = 4.08VPLTVNFDD2175 pKa = 3.92GSGSTQGDD2183 pKa = 3.8NPIVSYY2189 pKa = 10.88AWDD2192 pKa = 4.03FGDD2195 pKa = 4.81GNTSGIAEE2203 pKa = 4.32PVHH2206 pKa = 6.9EE2207 pKa = 4.28YY2208 pKa = 8.34TAPGVYY2214 pKa = 8.91TAVLTVVDD2222 pKa = 4.27EE2223 pKa = 4.97LGLEE2227 pKa = 4.18STDD2230 pKa = 3.64EE2231 pKa = 4.21IEE2233 pKa = 3.96ITVNDD2238 pKa = 4.06IVPEE2242 pKa = 4.21GDD2244 pKa = 3.25AVISFTLINADD2255 pKa = 3.15TDD2257 pKa = 3.72ADD2259 pKa = 4.24LLILSEE2265 pKa = 4.59GIIVDD2270 pKa = 3.92ATAVNGTGLNIRR2282 pKa = 11.84ANTNPLQVGSVVLQLTGPVGNLRR2305 pKa = 11.84TEE2307 pKa = 4.23SVAPYY2312 pKa = 11.06ALFGDD2317 pKa = 3.64SGGNYY2322 pKa = 9.68FGQALPEE2329 pKa = 4.19GNYY2332 pKa = 9.95TITATPFSASGGGGTPGTPLTINFSIGDD2360 pKa = 3.77PVIVVGPTAIATSNLQSGEE2379 pKa = 4.14APLEE2383 pKa = 3.76ILFNGSGSLEE2393 pKa = 3.99GDD2395 pKa = 3.38NPIINYY2401 pKa = 9.69AWDD2404 pKa = 4.05FGDD2407 pKa = 4.56GGSSEE2412 pKa = 4.67QPDD2415 pKa = 3.44PTYY2418 pKa = 11.27NYY2420 pKa = 7.37NTPGNYY2426 pKa = 7.74TATLTVTDD2434 pKa = 4.44DD2435 pKa = 4.52QGLQDD2440 pKa = 4.32SDD2442 pKa = 3.57QLIISVSGPVITGPTAIAVSDD2463 pKa = 4.01VTSGEE2468 pKa = 4.06APFEE2472 pKa = 4.68INFNGSGSVQGDD2484 pKa = 3.68NPITTYY2490 pKa = 10.66SWDD2493 pKa = 3.7FGDD2496 pKa = 5.59GGTSDD2501 pKa = 5.12LADD2504 pKa = 3.42PTYY2507 pKa = 10.76TYY2509 pKa = 8.17LTPGSFLAVLTVTDD2523 pKa = 4.25DD2524 pKa = 3.75QGLQATDD2531 pKa = 4.2EE2532 pKa = 4.33INITVTPPAPEE2543 pKa = 4.64ADD2545 pKa = 3.27QVVSFTLINAATDD2558 pKa = 3.28NDD2560 pKa = 4.24LFDD2563 pKa = 3.52ITEE2566 pKa = 4.62GIIIDD2571 pKa = 4.35LNSIGTPSLNIRR2583 pKa = 11.84ANTSPAVVGSVIMQLSGPVSNSRR2606 pKa = 11.84TEE2608 pKa = 4.09SVAPYY2613 pKa = 10.91ALFGDD2618 pKa = 3.89SSGNYY2623 pKa = 9.11FNGSLTPGDD2632 pKa = 4.09YY2633 pKa = 11.01SLTATPYY2640 pKa = 8.36TAGGGGGTAGTPLVINFSVVNTQTKK2665 pKa = 10.09SGTIAPDD2672 pKa = 3.11SSLGEE2677 pKa = 3.95EE2678 pKa = 4.26TVLSEE2683 pKa = 5.19EE2684 pKa = 4.23VLLYY2688 pKa = 9.5PNPTNYY2694 pKa = 9.46TLNISGLSSTDD2705 pKa = 2.67ISQEE2709 pKa = 3.35ARR2711 pKa = 11.84FYY2713 pKa = 10.04MHH2715 pKa = 7.68DD2716 pKa = 3.21IKK2718 pKa = 11.24GALLKK2723 pKa = 10.03TFTPNGVEE2731 pKa = 4.18SADD2734 pKa = 3.94GYY2736 pKa = 11.45ALPVDD2741 pKa = 4.83DD2742 pKa = 5.14LQQGVYY2748 pKa = 10.51LLSLEE2753 pKa = 4.4SNGRR2757 pKa = 11.84VLLRR2761 pKa = 11.84KK2762 pKa = 9.59RR2763 pKa = 11.84FVVMKK2768 pKa = 9.76QQ2769 pKa = 2.79
Molecular weight: 292.27 kDa
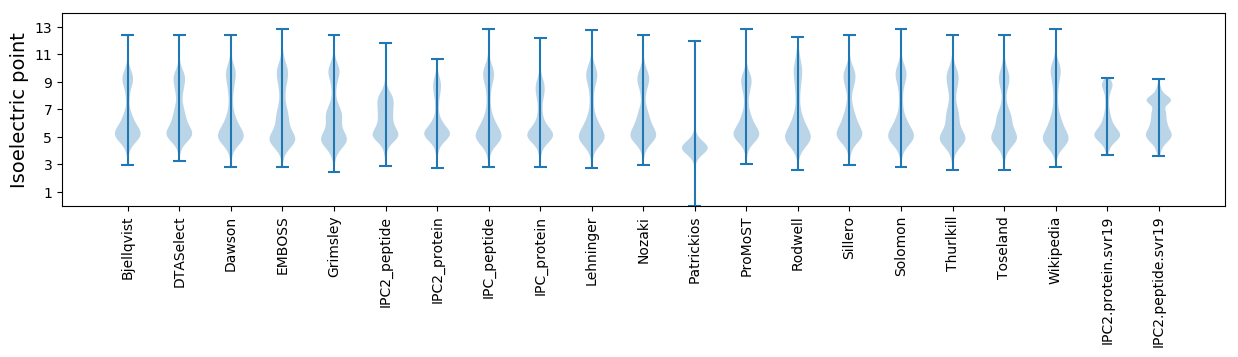
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S3LXR8|A0A4S3LXR8_9FLAO Membrane or secreted protein OS=Robertkochia marina OX=1227945 GN=E7Z59_11125 PE=4 SV=1
MM1 pKa = 7.3KK2 pKa = 10.2RR3 pKa = 11.84ISSVIASFFIVLLLVLAVSSCANARR28 pKa = 11.84WGTSAGVDD36 pKa = 3.96VVWGPGGPRR45 pKa = 11.84VQPNINVGVYY55 pKa = 10.36NGGRR59 pKa = 11.84WW60 pKa = 3.09
MM1 pKa = 7.3KK2 pKa = 10.2RR3 pKa = 11.84ISSVIASFFIVLLLVLAVSSCANARR28 pKa = 11.84WGTSAGVDD36 pKa = 3.96VVWGPGGPRR45 pKa = 11.84VQPNINVGVYY55 pKa = 10.36NGGRR59 pKa = 11.84WW60 pKa = 3.09
Molecular weight: 6.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1049829 |
38 |
4104 |
353.0 |
39.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.955 ± 0.045 | 0.725 ± 0.014 |
5.712 ± 0.037 | 7.206 ± 0.04 |
4.931 ± 0.032 | 7.006 ± 0.044 |
1.965 ± 0.024 | 6.917 ± 0.044 |
6.506 ± 0.052 | 9.612 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.024 | 5.221 ± 0.046 |
3.885 ± 0.025 | 3.404 ± 0.023 |
4.263 ± 0.031 | 6.12 ± 0.034 |
5.504 ± 0.042 | 6.423 ± 0.039 |
1.17 ± 0.016 | 3.998 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
