
Salix brachista
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Salix
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
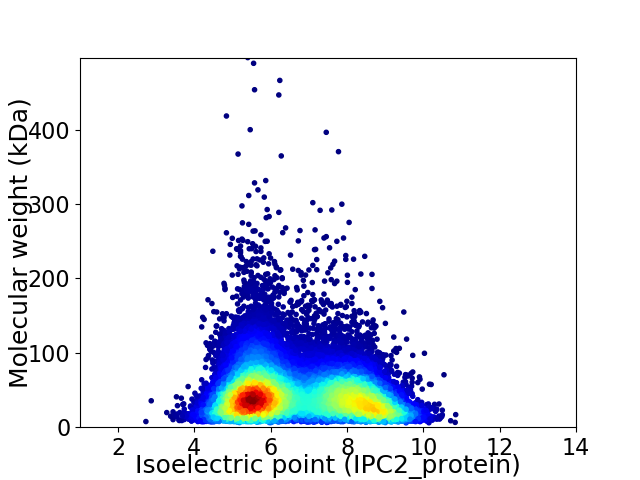
Virtual 2D-PAGE plot for 29803 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A5N5L4Z0|A0A5N5L4Z0_9ROSI Lipoxygenase domain-containing protein OS=Salix brachista OX=2182728 GN=DKX38_015350 PE=4 SV=1
MM1 pKa = 8.43IMEE4 pKa = 4.09VHH6 pKa = 6.37EE7 pKa = 4.73MLFCAPKK14 pKa = 8.97IYY16 pKa = 9.98FYY18 pKa = 11.22EE19 pKa = 4.64FVHH22 pKa = 6.03PQFGGIGCGDD32 pKa = 4.13LEE34 pKa = 4.54EE35 pKa = 4.38VWVTILNITGPLSSWSFADD54 pKa = 5.0NILPDD59 pKa = 3.94PEE61 pKa = 4.48IVDD64 pKa = 4.35GGPPSYY70 pKa = 10.11MLRR73 pKa = 11.84LSGTTQANWTFWLEE87 pKa = 3.96ASSSDD92 pKa = 3.66DD93 pKa = 3.26LRR95 pKa = 11.84VEE97 pKa = 4.23VAVVDD102 pKa = 3.84QVLDD106 pKa = 4.0DD107 pKa = 3.72EE108 pKa = 4.77VEE110 pKa = 4.04RR111 pKa = 11.84LKK113 pKa = 11.45GLFPDD118 pKa = 3.54WADD121 pKa = 3.06VATYY125 pKa = 10.66SSFMSSYY132 pKa = 11.3NFF134 pKa = 3.46
MM1 pKa = 8.43IMEE4 pKa = 4.09VHH6 pKa = 6.37EE7 pKa = 4.73MLFCAPKK14 pKa = 8.97IYY16 pKa = 9.98FYY18 pKa = 11.22EE19 pKa = 4.64FVHH22 pKa = 6.03PQFGGIGCGDD32 pKa = 4.13LEE34 pKa = 4.54EE35 pKa = 4.38VWVTILNITGPLSSWSFADD54 pKa = 5.0NILPDD59 pKa = 3.94PEE61 pKa = 4.48IVDD64 pKa = 4.35GGPPSYY70 pKa = 10.11MLRR73 pKa = 11.84LSGTTQANWTFWLEE87 pKa = 3.96ASSSDD92 pKa = 3.66DD93 pKa = 3.26LRR95 pKa = 11.84VEE97 pKa = 4.23VAVVDD102 pKa = 3.84QVLDD106 pKa = 4.0DD107 pKa = 3.72EE108 pKa = 4.77VEE110 pKa = 4.04RR111 pKa = 11.84LKK113 pKa = 11.45GLFPDD118 pKa = 3.54WADD121 pKa = 3.06VATYY125 pKa = 10.66SSFMSSYY132 pKa = 11.3NFF134 pKa = 3.46
Molecular weight: 15.13 kDa
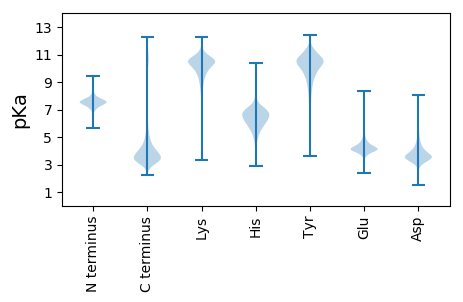
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N5LYA9|A0A5N5LYA9_9ROSI Uncharacterized protein OS=Salix brachista OX=2182728 GN=DKX38_011038 PE=4 SV=1
MM1 pKa = 7.36GSVVSQAANGIGGVVGNAFAVPIKK25 pKa = 9.75TLFGVSCEE33 pKa = 4.49DD34 pKa = 3.57VCSGPWDD41 pKa = 4.44LICFIEE47 pKa = 4.31HH48 pKa = 6.63LCVSDD53 pKa = 4.26LLKK56 pKa = 11.2LLMIFALSYY65 pKa = 8.15LTLMFFYY72 pKa = 11.16LLFKK76 pKa = 11.01VGICQCIGKK85 pKa = 8.45SLCKK89 pKa = 10.02ICWAGCEE96 pKa = 4.32AYY98 pKa = 9.19WFALEE103 pKa = 6.28DD104 pKa = 3.33ITCFLWHH111 pKa = 6.88KK112 pKa = 10.26LKK114 pKa = 9.24NTKK117 pKa = 9.64RR118 pKa = 11.84VNRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84FRR126 pKa = 11.84DD127 pKa = 2.84IEE129 pKa = 4.14GGYY132 pKa = 8.69TSSSEE137 pKa = 4.08SDD139 pKa = 3.22SSEE142 pKa = 4.43DD143 pKa = 3.44YY144 pKa = 10.91HH145 pKa = 8.77HH146 pKa = 7.47LGRR149 pKa = 11.84KK150 pKa = 8.5NKK152 pKa = 10.36SGMEE156 pKa = 3.85RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84TRR161 pKa = 11.84SRR163 pKa = 11.84SWRR166 pKa = 11.84SLHH169 pKa = 5.88PSSRR173 pKa = 11.84FGSRR177 pKa = 11.84NNHH180 pKa = 5.61HH181 pKa = 6.36RR182 pKa = 11.84HH183 pKa = 4.84HH184 pKa = 6.72VRR186 pKa = 11.84LKK188 pKa = 7.81TRR190 pKa = 11.84EE191 pKa = 3.7ISVHH195 pKa = 4.88VKK197 pKa = 9.95GGSRR201 pKa = 11.84RR202 pKa = 11.84PRR204 pKa = 11.84NSRR207 pKa = 11.84QLQLRR212 pKa = 11.84LRR214 pKa = 11.84NPRR217 pKa = 11.84RR218 pKa = 11.84NMGMFKK224 pKa = 10.38RR225 pKa = 11.84KK226 pKa = 9.6RR227 pKa = 11.84LGG229 pKa = 3.18
MM1 pKa = 7.36GSVVSQAANGIGGVVGNAFAVPIKK25 pKa = 9.75TLFGVSCEE33 pKa = 4.49DD34 pKa = 3.57VCSGPWDD41 pKa = 4.44LICFIEE47 pKa = 4.31HH48 pKa = 6.63LCVSDD53 pKa = 4.26LLKK56 pKa = 11.2LLMIFALSYY65 pKa = 8.15LTLMFFYY72 pKa = 11.16LLFKK76 pKa = 11.01VGICQCIGKK85 pKa = 8.45SLCKK89 pKa = 10.02ICWAGCEE96 pKa = 4.32AYY98 pKa = 9.19WFALEE103 pKa = 6.28DD104 pKa = 3.33ITCFLWHH111 pKa = 6.88KK112 pKa = 10.26LKK114 pKa = 9.24NTKK117 pKa = 9.64RR118 pKa = 11.84VNRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84FRR126 pKa = 11.84DD127 pKa = 2.84IEE129 pKa = 4.14GGYY132 pKa = 8.69TSSSEE137 pKa = 4.08SDD139 pKa = 3.22SSEE142 pKa = 4.43DD143 pKa = 3.44YY144 pKa = 10.91HH145 pKa = 8.77HH146 pKa = 7.47LGRR149 pKa = 11.84KK150 pKa = 8.5NKK152 pKa = 10.36SGMEE156 pKa = 3.85RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84TRR161 pKa = 11.84SRR163 pKa = 11.84SWRR166 pKa = 11.84SLHH169 pKa = 5.88PSSRR173 pKa = 11.84FGSRR177 pKa = 11.84NNHH180 pKa = 5.61HH181 pKa = 6.36RR182 pKa = 11.84HH183 pKa = 4.84HH184 pKa = 6.72VRR186 pKa = 11.84LKK188 pKa = 7.81TRR190 pKa = 11.84EE191 pKa = 3.7ISVHH195 pKa = 4.88VKK197 pKa = 9.95GGSRR201 pKa = 11.84RR202 pKa = 11.84PRR204 pKa = 11.84NSRR207 pKa = 11.84QLQLRR212 pKa = 11.84LRR214 pKa = 11.84NPRR217 pKa = 11.84RR218 pKa = 11.84NMGMFKK224 pKa = 10.38RR225 pKa = 11.84KK226 pKa = 9.6RR227 pKa = 11.84LGG229 pKa = 3.18
Molecular weight: 26.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13053624 |
51 |
5590 |
438.0 |
48.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.727 ± 0.012 | 1.924 ± 0.006 |
5.261 ± 0.009 | 6.44 ± 0.017 |
4.28 ± 0.009 | 6.593 ± 0.011 |
2.378 ± 0.005 | 5.261 ± 0.009 |
6.066 ± 0.014 | 9.784 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.005 | 4.431 ± 0.008 |
4.9 ± 0.012 | 3.69 ± 0.009 |
5.257 ± 0.01 | 9.167 ± 0.016 |
4.846 ± 0.009 | 6.494 ± 0.01 |
1.289 ± 0.005 | 2.746 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
