
Synechococcus phage S-CBS3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
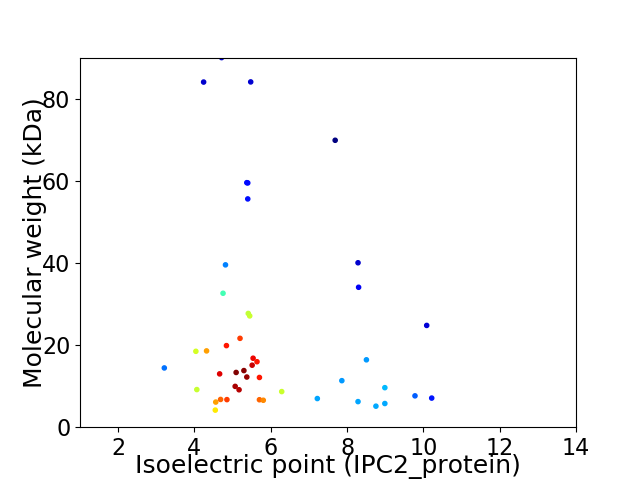
Virtual 2D-PAGE plot for 46 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4YCT0|F4YCT0_9CAUD Uncharacterized protein OS=Synechococcus phage S-CBS3 OX=753085 GN=S-CBS3_gp17 PE=4 SV=1
MM1 pKa = 7.53SLNISDD7 pKa = 4.75PLSLLAYY14 pKa = 10.09QSGLADD20 pKa = 3.98PPLVEE25 pKa = 5.11AAAQAADD32 pKa = 4.85DD33 pKa = 3.97LTAQQRR39 pKa = 11.84AYY41 pKa = 10.02KK42 pKa = 10.19IGDD45 pKa = 3.4PVPIVFCRR53 pKa = 11.84RR54 pKa = 11.84VSGNGGVMVAPGATEE69 pKa = 4.3ARR71 pKa = 11.84YY72 pKa = 10.14QNDD75 pKa = 3.42GTTNALTVSTMVVLSEE91 pKa = 4.67GEE93 pKa = 4.03LPQIDD98 pKa = 4.6LADD101 pKa = 4.16CFVGPCRR108 pKa = 11.84QGTWNQTYY116 pKa = 10.47DD117 pKa = 3.39RR118 pKa = 11.84RR119 pKa = 11.84AGTWTPGNFVTTVADD134 pKa = 3.9KK135 pKa = 11.19DD136 pKa = 3.72PWSCPYY142 pKa = 10.85YY143 pKa = 10.36CGTSGRR149 pKa = 11.84YY150 pKa = 8.93EE151 pKa = 4.65DD152 pKa = 4.48MTTMSCVNTFVDD164 pKa = 5.23GSEE167 pKa = 3.88RR168 pKa = 11.84WEE170 pKa = 4.19HH171 pKa = 4.92QLHH174 pKa = 5.17VFVRR178 pKa = 11.84EE179 pKa = 4.89GIQITRR185 pKa = 11.84IIDD188 pKa = 3.63STLGPSNNVIDD199 pKa = 4.17LALYY203 pKa = 11.15LMDD206 pKa = 3.96KK207 pKa = 10.28SGRR210 pKa = 11.84IPSTLIDD217 pKa = 3.86STQMLAAANFTEE229 pKa = 4.54TNGLHH234 pKa = 6.16FNGVFQEE241 pKa = 4.1SLNLDD246 pKa = 3.16EE247 pKa = 4.86WLEE250 pKa = 4.12QISNDD255 pKa = 3.26YY256 pKa = 10.91LLRR259 pKa = 11.84LVEE262 pKa = 4.33LNGKK266 pKa = 9.73FGFRR270 pKa = 11.84PRR272 pKa = 11.84LPVNVDD278 pKa = 2.96HH279 pKa = 7.06TIKK282 pKa = 8.14TTAIDD287 pKa = 3.07WDD289 pKa = 4.02YY290 pKa = 11.2TFTEE294 pKa = 4.39DD295 pKa = 5.18HH296 pKa = 6.81LLPDD300 pKa = 4.08GFEE303 pKa = 3.71IQYY306 pKa = 10.4IPLADD311 pKa = 5.69RR312 pKa = 11.84IPVTLQMMWRR322 pKa = 11.84QQPEE326 pKa = 3.8SDD328 pKa = 2.83IGFPRR333 pKa = 11.84TTEE336 pKa = 3.33ISYY339 pKa = 10.68SGEE342 pKa = 3.8AADD345 pKa = 6.44GPFEE349 pKa = 4.29QYY351 pKa = 10.91DD352 pKa = 3.56LSGYY356 pKa = 7.51CTSEE360 pKa = 3.57THH362 pKa = 5.58AVKK365 pKa = 10.75VGAYY369 pKa = 9.28RR370 pKa = 11.84LARR373 pKa = 11.84RR374 pKa = 11.84KK375 pKa = 9.96FITHH379 pKa = 5.66TLRR382 pKa = 11.84LKK384 pKa = 10.44VRR386 pKa = 11.84PANYY390 pKa = 10.26NSVLTQGDD398 pKa = 3.62IVRR401 pKa = 11.84VRR403 pKa = 11.84LRR405 pKa = 11.84RR406 pKa = 11.84EE407 pKa = 3.49TALAALDD414 pKa = 3.76YY415 pKa = 11.16HH416 pKa = 6.99DD417 pKa = 5.13FLYY420 pKa = 10.71EE421 pKa = 3.55IEE423 pKa = 5.16RR424 pKa = 11.84IEE426 pKa = 4.09KK427 pKa = 8.06TASGACIFDD436 pKa = 3.79LTHH439 pKa = 6.8FPIDD443 pKa = 3.4AQGRR447 pKa = 11.84SLVALEE453 pKa = 4.29VAAATAPGVVIASGRR468 pKa = 11.84SDD470 pKa = 3.7YY471 pKa = 11.72SCDD474 pKa = 4.17DD475 pKa = 3.56NSSTPGTSVGSGGIDD490 pKa = 3.51FPAFDD495 pKa = 5.0DD496 pKa = 4.54TPGLDD501 pKa = 3.52DD502 pKa = 3.54TTVDD506 pKa = 4.35FNVPTDD512 pKa = 4.28DD513 pKa = 3.19EE514 pKa = 4.68WATGGYY520 pKa = 9.73SPIGPDD526 pKa = 3.13VSQPAGEE533 pKa = 4.14PSGGQTPIGGWDD545 pKa = 3.53NPADD549 pKa = 4.12PLEE552 pKa = 4.46STLDD556 pKa = 3.58QSGVGYY562 pKa = 8.54LTGGTGTGGAPQSGDD577 pKa = 3.51TLSVADD583 pKa = 4.66TDD585 pKa = 4.38FTCAGQVCWSKK596 pKa = 10.82INKK599 pKa = 7.82DD600 pKa = 3.15TGVQTDD606 pKa = 4.01ISCQDD611 pKa = 3.47EE612 pKa = 4.97PISGSWDD619 pKa = 3.27LSITTDD625 pKa = 3.47EE626 pKa = 3.96VDD628 pKa = 3.4HH629 pKa = 6.73FIVATGRR636 pKa = 11.84CKK638 pKa = 10.87DD639 pKa = 3.39PSTASGWGPPQSLGQTAAVLPPAVPSCGDD668 pKa = 3.57LVSVTIKK675 pKa = 10.11WKK677 pKa = 10.0MSIAGYY683 pKa = 8.94TSACVPSTSICSAFGDD699 pKa = 3.74NQYY702 pKa = 11.36SKK704 pKa = 9.82TITSVPRR711 pKa = 11.84GTQIDD716 pKa = 3.72FGGQINAGAGTCADD730 pKa = 4.93KK731 pKa = 11.09INAAISYY738 pKa = 6.97WKK740 pKa = 10.27CIGGVATYY748 pKa = 8.53VTEE751 pKa = 4.3ALGTPGCSYY760 pKa = 10.84NVSAGYY766 pKa = 7.74TQTPYY771 pKa = 10.89DD772 pKa = 3.99YY773 pKa = 10.55EE774 pKa = 4.2AVGDD778 pKa = 3.86VTYY781 pKa = 10.12TGPP784 pKa = 3.79
MM1 pKa = 7.53SLNISDD7 pKa = 4.75PLSLLAYY14 pKa = 10.09QSGLADD20 pKa = 3.98PPLVEE25 pKa = 5.11AAAQAADD32 pKa = 4.85DD33 pKa = 3.97LTAQQRR39 pKa = 11.84AYY41 pKa = 10.02KK42 pKa = 10.19IGDD45 pKa = 3.4PVPIVFCRR53 pKa = 11.84RR54 pKa = 11.84VSGNGGVMVAPGATEE69 pKa = 4.3ARR71 pKa = 11.84YY72 pKa = 10.14QNDD75 pKa = 3.42GTTNALTVSTMVVLSEE91 pKa = 4.67GEE93 pKa = 4.03LPQIDD98 pKa = 4.6LADD101 pKa = 4.16CFVGPCRR108 pKa = 11.84QGTWNQTYY116 pKa = 10.47DD117 pKa = 3.39RR118 pKa = 11.84RR119 pKa = 11.84AGTWTPGNFVTTVADD134 pKa = 3.9KK135 pKa = 11.19DD136 pKa = 3.72PWSCPYY142 pKa = 10.85YY143 pKa = 10.36CGTSGRR149 pKa = 11.84YY150 pKa = 8.93EE151 pKa = 4.65DD152 pKa = 4.48MTTMSCVNTFVDD164 pKa = 5.23GSEE167 pKa = 3.88RR168 pKa = 11.84WEE170 pKa = 4.19HH171 pKa = 4.92QLHH174 pKa = 5.17VFVRR178 pKa = 11.84EE179 pKa = 4.89GIQITRR185 pKa = 11.84IIDD188 pKa = 3.63STLGPSNNVIDD199 pKa = 4.17LALYY203 pKa = 11.15LMDD206 pKa = 3.96KK207 pKa = 10.28SGRR210 pKa = 11.84IPSTLIDD217 pKa = 3.86STQMLAAANFTEE229 pKa = 4.54TNGLHH234 pKa = 6.16FNGVFQEE241 pKa = 4.1SLNLDD246 pKa = 3.16EE247 pKa = 4.86WLEE250 pKa = 4.12QISNDD255 pKa = 3.26YY256 pKa = 10.91LLRR259 pKa = 11.84LVEE262 pKa = 4.33LNGKK266 pKa = 9.73FGFRR270 pKa = 11.84PRR272 pKa = 11.84LPVNVDD278 pKa = 2.96HH279 pKa = 7.06TIKK282 pKa = 8.14TTAIDD287 pKa = 3.07WDD289 pKa = 4.02YY290 pKa = 11.2TFTEE294 pKa = 4.39DD295 pKa = 5.18HH296 pKa = 6.81LLPDD300 pKa = 4.08GFEE303 pKa = 3.71IQYY306 pKa = 10.4IPLADD311 pKa = 5.69RR312 pKa = 11.84IPVTLQMMWRR322 pKa = 11.84QQPEE326 pKa = 3.8SDD328 pKa = 2.83IGFPRR333 pKa = 11.84TTEE336 pKa = 3.33ISYY339 pKa = 10.68SGEE342 pKa = 3.8AADD345 pKa = 6.44GPFEE349 pKa = 4.29QYY351 pKa = 10.91DD352 pKa = 3.56LSGYY356 pKa = 7.51CTSEE360 pKa = 3.57THH362 pKa = 5.58AVKK365 pKa = 10.75VGAYY369 pKa = 9.28RR370 pKa = 11.84LARR373 pKa = 11.84RR374 pKa = 11.84KK375 pKa = 9.96FITHH379 pKa = 5.66TLRR382 pKa = 11.84LKK384 pKa = 10.44VRR386 pKa = 11.84PANYY390 pKa = 10.26NSVLTQGDD398 pKa = 3.62IVRR401 pKa = 11.84VRR403 pKa = 11.84LRR405 pKa = 11.84RR406 pKa = 11.84EE407 pKa = 3.49TALAALDD414 pKa = 3.76YY415 pKa = 11.16HH416 pKa = 6.99DD417 pKa = 5.13FLYY420 pKa = 10.71EE421 pKa = 3.55IEE423 pKa = 5.16RR424 pKa = 11.84IEE426 pKa = 4.09KK427 pKa = 8.06TASGACIFDD436 pKa = 3.79LTHH439 pKa = 6.8FPIDD443 pKa = 3.4AQGRR447 pKa = 11.84SLVALEE453 pKa = 4.29VAAATAPGVVIASGRR468 pKa = 11.84SDD470 pKa = 3.7YY471 pKa = 11.72SCDD474 pKa = 4.17DD475 pKa = 3.56NSSTPGTSVGSGGIDD490 pKa = 3.51FPAFDD495 pKa = 5.0DD496 pKa = 4.54TPGLDD501 pKa = 3.52DD502 pKa = 3.54TTVDD506 pKa = 4.35FNVPTDD512 pKa = 4.28DD513 pKa = 3.19EE514 pKa = 4.68WATGGYY520 pKa = 9.73SPIGPDD526 pKa = 3.13VSQPAGEE533 pKa = 4.14PSGGQTPIGGWDD545 pKa = 3.53NPADD549 pKa = 4.12PLEE552 pKa = 4.46STLDD556 pKa = 3.58QSGVGYY562 pKa = 8.54LTGGTGTGGAPQSGDD577 pKa = 3.51TLSVADD583 pKa = 4.66TDD585 pKa = 4.38FTCAGQVCWSKK596 pKa = 10.82INKK599 pKa = 7.82DD600 pKa = 3.15TGVQTDD606 pKa = 4.01ISCQDD611 pKa = 3.47EE612 pKa = 4.97PISGSWDD619 pKa = 3.27LSITTDD625 pKa = 3.47EE626 pKa = 3.96VDD628 pKa = 3.4HH629 pKa = 6.73FIVATGRR636 pKa = 11.84CKK638 pKa = 10.87DD639 pKa = 3.39PSTASGWGPPQSLGQTAAVLPPAVPSCGDD668 pKa = 3.57LVSVTIKK675 pKa = 10.11WKK677 pKa = 10.0MSIAGYY683 pKa = 8.94TSACVPSTSICSAFGDD699 pKa = 3.74NQYY702 pKa = 11.36SKK704 pKa = 9.82TITSVPRR711 pKa = 11.84GTQIDD716 pKa = 3.72FGGQINAGAGTCADD730 pKa = 4.93KK731 pKa = 11.09INAAISYY738 pKa = 6.97WKK740 pKa = 10.27CIGGVATYY748 pKa = 8.53VTEE751 pKa = 4.3ALGTPGCSYY760 pKa = 10.84NVSAGYY766 pKa = 7.74TQTPYY771 pKa = 10.89DD772 pKa = 3.99YY773 pKa = 10.55EE774 pKa = 4.2AVGDD778 pKa = 3.86VTYY781 pKa = 10.12TGPP784 pKa = 3.79
Molecular weight: 84.18 kDa
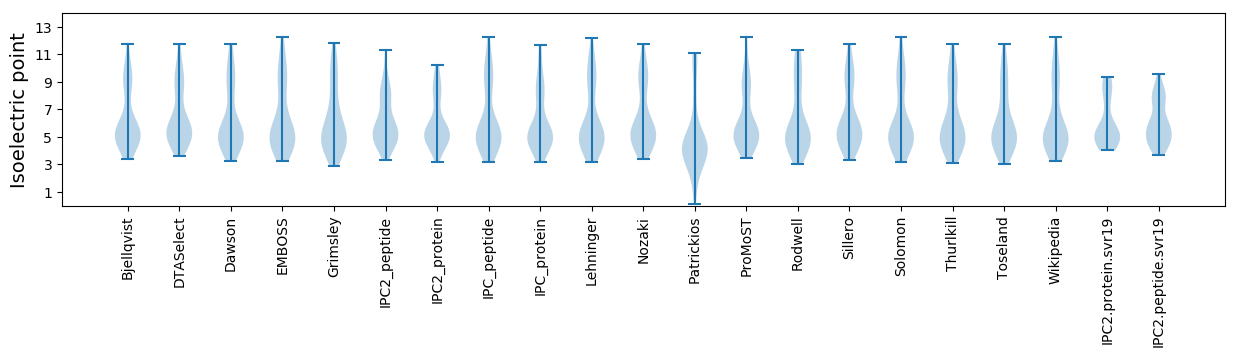
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4YCU6|F4YCU6_9CAUD Uncharacterized protein OS=Synechococcus phage S-CBS3 OX=753085 GN=S-CBS3_gp33 PE=4 SV=1
MM1 pKa = 7.75TDD3 pKa = 3.23PVLQEE8 pKa = 3.76LEE10 pKa = 3.87RR11 pKa = 11.84LRR13 pKa = 11.84EE14 pKa = 3.75YY15 pKa = 10.67RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ADD20 pKa = 2.96AKK22 pKa = 10.37RR23 pKa = 11.84RR24 pKa = 11.84DD25 pKa = 3.76RR26 pKa = 11.84QVQARR31 pKa = 11.84LPPKK35 pKa = 10.03LAQQFTAYY43 pKa = 8.64MQAHH47 pKa = 7.23GLNQNQAIVAILYY60 pKa = 7.61TFFNN64 pKa = 4.09
MM1 pKa = 7.75TDD3 pKa = 3.23PVLQEE8 pKa = 3.76LEE10 pKa = 3.87RR11 pKa = 11.84LRR13 pKa = 11.84EE14 pKa = 3.75YY15 pKa = 10.67RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ADD20 pKa = 2.96AKK22 pKa = 10.37RR23 pKa = 11.84RR24 pKa = 11.84DD25 pKa = 3.76RR26 pKa = 11.84QVQARR31 pKa = 11.84LPPKK35 pKa = 10.03LAQQFTAYY43 pKa = 8.64MQAHH47 pKa = 7.23GLNQNQAIVAILYY60 pKa = 7.61TFFNN64 pKa = 4.09
Molecular weight: 7.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9963 |
40 |
836 |
216.6 |
23.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.312 ± 0.479 | 1.174 ± 0.194 |
6.072 ± 0.253 | 5.37 ± 0.317 |
3.433 ± 0.259 | 8.301 ± 0.421 |
1.787 ± 0.211 | 5.099 ± 0.279 |
3.192 ± 0.253 | 8.15 ± 0.401 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.389 ± 0.201 | 2.901 ± 0.155 |
5.179 ± 0.312 | 4.487 ± 0.194 |
6.243 ± 0.405 | 6.635 ± 0.306 |
7.488 ± 0.525 | 6.213 ± 0.27 |
1.887 ± 0.172 | 2.69 ± 0.211 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
