
Quaranfil virus (isolate QrfV/Tick/Afghanistan/EG_T_377/1968) (QRFV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Quaranjavirus; Quaranfil quaranjavirus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
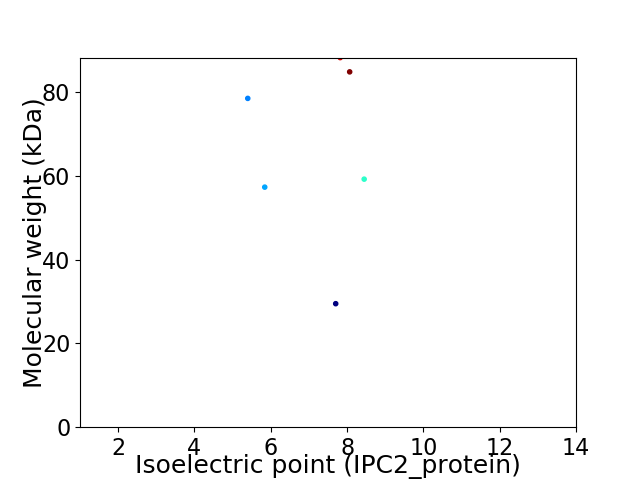
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|D0UFC9|M1_QRFVE Probable matrix protein OS=Quaranfil virus (isolate QrfV/Tick/Afghanistan/EG_T_377/1968) OX=1559362 GN=Segment 6 PE=4 SV=1
MM1 pKa = 7.4AFEE4 pKa = 5.3AYY6 pKa = 9.91RR7 pKa = 11.84LLIANPQLYY16 pKa = 9.98EE17 pKa = 4.13PDD19 pKa = 3.81FVSQAGEE26 pKa = 4.1LSEE29 pKa = 3.98HH30 pKa = 6.37WARR33 pKa = 11.84EE34 pKa = 3.45NWKK37 pKa = 10.16RR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 3.51EE41 pKa = 4.18SLRR44 pKa = 11.84HH45 pKa = 6.34DD46 pKa = 4.3KK47 pKa = 11.27VCMLLMNTEE56 pKa = 3.83PRR58 pKa = 11.84EE59 pKa = 4.05RR60 pKa = 11.84SHH62 pKa = 5.81TQAEE66 pKa = 4.66GEE68 pKa = 4.36EE69 pKa = 4.53EE70 pKa = 4.53GSSSRR75 pKa = 11.84SLATEE80 pKa = 3.78EE81 pKa = 4.42GEE83 pKa = 4.64KK84 pKa = 10.55RR85 pKa = 11.84FSLEE89 pKa = 3.77ALEE92 pKa = 4.69GLVDD96 pKa = 3.5QGRR99 pKa = 11.84EE100 pKa = 3.82VLLEE104 pKa = 3.92NLEE107 pKa = 4.15MEE109 pKa = 4.57EE110 pKa = 4.06AQANSDD116 pKa = 3.86SEE118 pKa = 4.69GPNEE122 pKa = 4.2ADD124 pKa = 2.97SGGEE128 pKa = 3.89GEE130 pKa = 4.3EE131 pKa = 4.09FVFGVEE137 pKa = 4.6SEE139 pKa = 4.72SEE141 pKa = 4.09MEE143 pKa = 4.3VASEE147 pKa = 4.15VTKK150 pKa = 10.9LLEE153 pKa = 4.74PDD155 pKa = 5.25LIDD158 pKa = 4.0SNFRR162 pKa = 11.84YY163 pKa = 8.19TLLEE167 pKa = 4.12GVTSGKK173 pKa = 9.1FAQMEE178 pKa = 4.5FTALWGINTNNKK190 pKa = 8.17WDD192 pKa = 3.61LVDD195 pKa = 3.5RR196 pKa = 11.84VNRR199 pKa = 11.84KK200 pKa = 9.48LIEE203 pKa = 4.32VKK205 pKa = 9.45VTTRR209 pKa = 11.84PPTGVWEE216 pKa = 4.22EE217 pKa = 4.34VVAHH221 pKa = 7.04AEE223 pKa = 4.31GTDD226 pKa = 3.65PEE228 pKa = 4.86HH229 pKa = 6.36NGAYY233 pKa = 9.54IIHH236 pKa = 7.96DD237 pKa = 4.15DD238 pKa = 3.63LCGNFTPYY246 pKa = 10.95KK247 pKa = 9.69FGNISDD253 pKa = 4.08LPGWPSAQDD262 pKa = 3.44FPIRR266 pKa = 11.84RR267 pKa = 11.84HH268 pKa = 5.93AFLASYY274 pKa = 10.62GGVPSAGGIEE284 pKa = 4.23EE285 pKa = 5.3GLPAWDD291 pKa = 3.82DD292 pKa = 3.38VFMEE296 pKa = 4.76RR297 pKa = 11.84TRR299 pKa = 11.84NWVAPLWKK307 pKa = 9.82KK308 pKa = 10.93GPISVRR314 pKa = 11.84RR315 pKa = 11.84DD316 pKa = 3.09DD317 pKa = 4.32CPRR320 pKa = 11.84MEE322 pKa = 5.16PFNITKK328 pKa = 10.18FITLLEE334 pKa = 4.32DD335 pKa = 3.33PRR337 pKa = 11.84LRR339 pKa = 11.84NTDD342 pKa = 4.12KK343 pKa = 10.54SAKK346 pKa = 8.36WKK348 pKa = 10.84GKK350 pKa = 9.1ILPVSWCRR358 pKa = 11.84TIISSQEE365 pKa = 3.31KK366 pKa = 10.37DD367 pKa = 2.41IDD369 pKa = 3.74MVQEE373 pKa = 4.12VIRR376 pKa = 11.84DD377 pKa = 3.62LGVIGLWEE385 pKa = 4.87EE386 pKa = 4.09INANPVKK393 pKa = 10.77VEE395 pKa = 4.01RR396 pKa = 11.84TVSALSEE403 pKa = 3.99LMRR406 pKa = 11.84LFQEE410 pKa = 4.94GFRR413 pKa = 11.84EE414 pKa = 4.07KK415 pKa = 10.88NPYY418 pKa = 9.77ISVKK422 pKa = 9.83RR423 pKa = 11.84KK424 pKa = 9.06CKK426 pKa = 8.78GWVITGHH433 pKa = 5.98KK434 pKa = 7.31QTPALTDD441 pKa = 3.24TLLEE445 pKa = 4.56LGVGYY450 pKa = 10.43KK451 pKa = 9.84PYY453 pKa = 10.87QMSNVVEE460 pKa = 3.97EE461 pKa = 5.15DD462 pKa = 3.06MRR464 pKa = 11.84QCEE467 pKa = 3.92TDD469 pKa = 4.68DD470 pKa = 3.52IEE472 pKa = 4.62KK473 pKa = 10.07MKK475 pKa = 10.33WVDD478 pKa = 3.15WMSKK482 pKa = 9.83LAATEE487 pKa = 4.15SEE489 pKa = 4.12PLGSKK494 pKa = 9.89VSRR497 pKa = 11.84DD498 pKa = 3.96DD499 pKa = 3.16IFKK502 pKa = 9.76EE503 pKa = 3.94AEE505 pKa = 3.95SRR507 pKa = 11.84HH508 pKa = 5.4VLDD511 pKa = 4.76EE512 pKa = 4.28PAKK515 pKa = 9.63KK516 pKa = 9.02MCSKK520 pKa = 10.86VYY522 pKa = 10.62DD523 pKa = 4.32LYY525 pKa = 11.0RR526 pKa = 11.84SHH528 pKa = 7.57KK529 pKa = 10.37IGATCSKK536 pKa = 10.72FMGFYY541 pKa = 10.58SRR543 pKa = 11.84MGGSYY548 pKa = 10.47LRR550 pKa = 11.84AIAGNSRR557 pKa = 11.84QHH559 pKa = 6.22SSLAIMPLYY568 pKa = 10.23YY569 pKa = 9.22KK570 pKa = 9.95TYY572 pKa = 10.48QEE574 pKa = 5.26DD575 pKa = 4.18GTSTRR580 pKa = 11.84HH581 pKa = 4.52LTGLVIRR588 pKa = 11.84GPHH591 pKa = 5.92HH592 pKa = 6.27VRR594 pKa = 11.84EE595 pKa = 4.37STDD598 pKa = 3.51TINLLIVEE606 pKa = 4.61KK607 pKa = 9.44TCLSRR612 pKa = 11.84RR613 pKa = 11.84EE614 pKa = 4.09AQEE617 pKa = 3.88RR618 pKa = 11.84LSGGALVNGVWWVRR632 pKa = 11.84KK633 pKa = 8.74NAIRR637 pKa = 11.84KK638 pKa = 9.11ADD640 pKa = 3.58PTYY643 pKa = 11.01LAFLHH648 pKa = 6.51NSLFVPTNFLGEE660 pKa = 4.22LVTTPKK666 pKa = 10.53HH667 pKa = 6.08FLRR670 pKa = 11.84QRR672 pKa = 11.84QPRR675 pKa = 11.84ILGRR679 pKa = 11.84NPVWQLWLLL688 pKa = 3.93
MM1 pKa = 7.4AFEE4 pKa = 5.3AYY6 pKa = 9.91RR7 pKa = 11.84LLIANPQLYY16 pKa = 9.98EE17 pKa = 4.13PDD19 pKa = 3.81FVSQAGEE26 pKa = 4.1LSEE29 pKa = 3.98HH30 pKa = 6.37WARR33 pKa = 11.84EE34 pKa = 3.45NWKK37 pKa = 10.16RR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 3.51EE41 pKa = 4.18SLRR44 pKa = 11.84HH45 pKa = 6.34DD46 pKa = 4.3KK47 pKa = 11.27VCMLLMNTEE56 pKa = 3.83PRR58 pKa = 11.84EE59 pKa = 4.05RR60 pKa = 11.84SHH62 pKa = 5.81TQAEE66 pKa = 4.66GEE68 pKa = 4.36EE69 pKa = 4.53EE70 pKa = 4.53GSSSRR75 pKa = 11.84SLATEE80 pKa = 3.78EE81 pKa = 4.42GEE83 pKa = 4.64KK84 pKa = 10.55RR85 pKa = 11.84FSLEE89 pKa = 3.77ALEE92 pKa = 4.69GLVDD96 pKa = 3.5QGRR99 pKa = 11.84EE100 pKa = 3.82VLLEE104 pKa = 3.92NLEE107 pKa = 4.15MEE109 pKa = 4.57EE110 pKa = 4.06AQANSDD116 pKa = 3.86SEE118 pKa = 4.69GPNEE122 pKa = 4.2ADD124 pKa = 2.97SGGEE128 pKa = 3.89GEE130 pKa = 4.3EE131 pKa = 4.09FVFGVEE137 pKa = 4.6SEE139 pKa = 4.72SEE141 pKa = 4.09MEE143 pKa = 4.3VASEE147 pKa = 4.15VTKK150 pKa = 10.9LLEE153 pKa = 4.74PDD155 pKa = 5.25LIDD158 pKa = 4.0SNFRR162 pKa = 11.84YY163 pKa = 8.19TLLEE167 pKa = 4.12GVTSGKK173 pKa = 9.1FAQMEE178 pKa = 4.5FTALWGINTNNKK190 pKa = 8.17WDD192 pKa = 3.61LVDD195 pKa = 3.5RR196 pKa = 11.84VNRR199 pKa = 11.84KK200 pKa = 9.48LIEE203 pKa = 4.32VKK205 pKa = 9.45VTTRR209 pKa = 11.84PPTGVWEE216 pKa = 4.22EE217 pKa = 4.34VVAHH221 pKa = 7.04AEE223 pKa = 4.31GTDD226 pKa = 3.65PEE228 pKa = 4.86HH229 pKa = 6.36NGAYY233 pKa = 9.54IIHH236 pKa = 7.96DD237 pKa = 4.15DD238 pKa = 3.63LCGNFTPYY246 pKa = 10.95KK247 pKa = 9.69FGNISDD253 pKa = 4.08LPGWPSAQDD262 pKa = 3.44FPIRR266 pKa = 11.84RR267 pKa = 11.84HH268 pKa = 5.93AFLASYY274 pKa = 10.62GGVPSAGGIEE284 pKa = 4.23EE285 pKa = 5.3GLPAWDD291 pKa = 3.82DD292 pKa = 3.38VFMEE296 pKa = 4.76RR297 pKa = 11.84TRR299 pKa = 11.84NWVAPLWKK307 pKa = 9.82KK308 pKa = 10.93GPISVRR314 pKa = 11.84RR315 pKa = 11.84DD316 pKa = 3.09DD317 pKa = 4.32CPRR320 pKa = 11.84MEE322 pKa = 5.16PFNITKK328 pKa = 10.18FITLLEE334 pKa = 4.32DD335 pKa = 3.33PRR337 pKa = 11.84LRR339 pKa = 11.84NTDD342 pKa = 4.12KK343 pKa = 10.54SAKK346 pKa = 8.36WKK348 pKa = 10.84GKK350 pKa = 9.1ILPVSWCRR358 pKa = 11.84TIISSQEE365 pKa = 3.31KK366 pKa = 10.37DD367 pKa = 2.41IDD369 pKa = 3.74MVQEE373 pKa = 4.12VIRR376 pKa = 11.84DD377 pKa = 3.62LGVIGLWEE385 pKa = 4.87EE386 pKa = 4.09INANPVKK393 pKa = 10.77VEE395 pKa = 4.01RR396 pKa = 11.84TVSALSEE403 pKa = 3.99LMRR406 pKa = 11.84LFQEE410 pKa = 4.94GFRR413 pKa = 11.84EE414 pKa = 4.07KK415 pKa = 10.88NPYY418 pKa = 9.77ISVKK422 pKa = 9.83RR423 pKa = 11.84KK424 pKa = 9.06CKK426 pKa = 8.78GWVITGHH433 pKa = 5.98KK434 pKa = 7.31QTPALTDD441 pKa = 3.24TLLEE445 pKa = 4.56LGVGYY450 pKa = 10.43KK451 pKa = 9.84PYY453 pKa = 10.87QMSNVVEE460 pKa = 3.97EE461 pKa = 5.15DD462 pKa = 3.06MRR464 pKa = 11.84QCEE467 pKa = 3.92TDD469 pKa = 4.68DD470 pKa = 3.52IEE472 pKa = 4.62KK473 pKa = 10.07MKK475 pKa = 10.33WVDD478 pKa = 3.15WMSKK482 pKa = 9.83LAATEE487 pKa = 4.15SEE489 pKa = 4.12PLGSKK494 pKa = 9.89VSRR497 pKa = 11.84DD498 pKa = 3.96DD499 pKa = 3.16IFKK502 pKa = 9.76EE503 pKa = 3.94AEE505 pKa = 3.95SRR507 pKa = 11.84HH508 pKa = 5.4VLDD511 pKa = 4.76EE512 pKa = 4.28PAKK515 pKa = 9.63KK516 pKa = 9.02MCSKK520 pKa = 10.86VYY522 pKa = 10.62DD523 pKa = 4.32LYY525 pKa = 11.0RR526 pKa = 11.84SHH528 pKa = 7.57KK529 pKa = 10.37IGATCSKK536 pKa = 10.72FMGFYY541 pKa = 10.58SRR543 pKa = 11.84MGGSYY548 pKa = 10.47LRR550 pKa = 11.84AIAGNSRR557 pKa = 11.84QHH559 pKa = 6.22SSLAIMPLYY568 pKa = 10.23YY569 pKa = 9.22KK570 pKa = 9.95TYY572 pKa = 10.48QEE574 pKa = 5.26DD575 pKa = 4.18GTSTRR580 pKa = 11.84HH581 pKa = 4.52LTGLVIRR588 pKa = 11.84GPHH591 pKa = 5.92HH592 pKa = 6.27VRR594 pKa = 11.84EE595 pKa = 4.37STDD598 pKa = 3.51TINLLIVEE606 pKa = 4.61KK607 pKa = 9.44TCLSRR612 pKa = 11.84RR613 pKa = 11.84EE614 pKa = 4.09AQEE617 pKa = 3.88RR618 pKa = 11.84LSGGALVNGVWWVRR632 pKa = 11.84KK633 pKa = 8.74NAIRR637 pKa = 11.84KK638 pKa = 9.11ADD640 pKa = 3.58PTYY643 pKa = 11.01LAFLHH648 pKa = 6.51NSLFVPTNFLGEE660 pKa = 4.22LVTTPKK666 pKa = 10.53HH667 pKa = 6.08FLRR670 pKa = 11.84QRR672 pKa = 11.84QPRR675 pKa = 11.84ILGRR679 pKa = 11.84NPVWQLWLLL688 pKa = 3.93
Molecular weight: 78.46 kDa
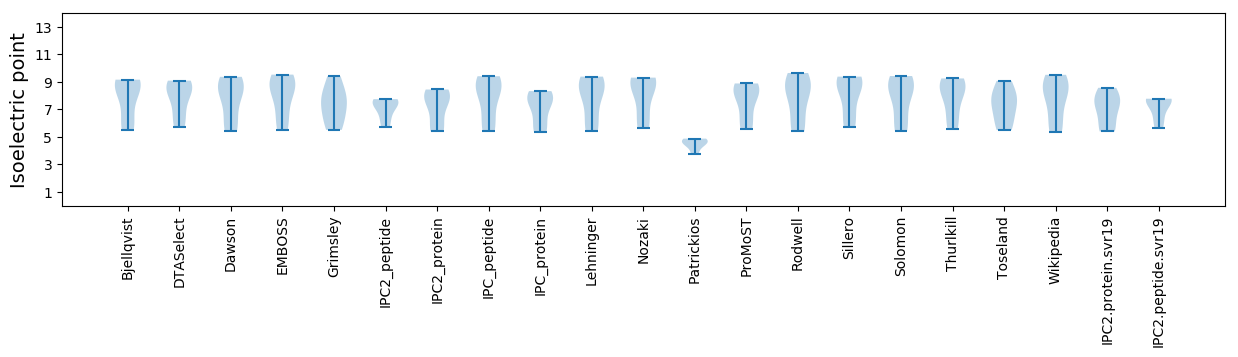
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|G9HXM2|NCAP_QRFVE Nucleoprotein OS=Quaranfil virus (isolate QrfV/Tick/Afghanistan/EG_T_377/1968) OX=1559362 GN=Segment 4 PE=3 SV=1
MM1 pKa = 7.43EE2 pKa = 5.29VDD4 pKa = 4.5PAPSTAPKK12 pKa = 8.41PTRR15 pKa = 11.84KK16 pKa = 9.36RR17 pKa = 11.84VNNSGEE23 pKa = 4.21EE24 pKa = 3.82IGSGGKK30 pKa = 9.41RR31 pKa = 11.84ALTGLSLTQKK41 pKa = 10.2VAIHH45 pKa = 5.83HH46 pKa = 7.14AIALMVYY53 pKa = 9.66HH54 pKa = 5.99VVIDD58 pKa = 3.62IFKK61 pKa = 9.33CTIIDD66 pKa = 5.58LITDD70 pKa = 3.48VNIGVYY76 pKa = 10.71AMNQIWSIHH85 pKa = 5.46NYY87 pKa = 9.26FRR89 pKa = 11.84TKK91 pKa = 10.61NNSGRR96 pKa = 11.84SSIGAKK102 pKa = 9.28IDD104 pKa = 3.55NATDD108 pKa = 3.75TYY110 pKa = 11.41QFILHH115 pKa = 6.34GRR117 pKa = 11.84TITVNKK123 pKa = 10.06EE124 pKa = 3.72EE125 pKa = 4.33LKK127 pKa = 10.8GVYY130 pKa = 8.98KK131 pKa = 10.59RR132 pKa = 11.84CITTYY137 pKa = 10.83FDD139 pKa = 3.42WDD141 pKa = 4.3KK142 pKa = 11.12MNDD145 pKa = 3.09WMGSVHH151 pKa = 7.41TIMALWNLFGSRR163 pKa = 11.84LSEE166 pKa = 3.88VRR168 pKa = 11.84IMPSANTISVEE179 pKa = 3.77KK180 pKa = 10.59DD181 pKa = 2.71NRR183 pKa = 11.84TTVVKK188 pKa = 10.68DD189 pKa = 3.62FNQYY193 pKa = 10.48GIPAGMRR200 pKa = 11.84HH201 pKa = 5.99FATGADD207 pKa = 4.53FKK209 pKa = 10.69PTMKK213 pKa = 10.48SALAQSMGPVTVLVQLSEE231 pKa = 4.6AKK233 pKa = 10.45DD234 pKa = 3.39NQYY237 pKa = 11.0AGKK240 pKa = 9.27WVDD243 pKa = 3.04ALKK246 pKa = 10.54RR247 pKa = 11.84AFSHH251 pKa = 6.57IPHH254 pKa = 7.38IDD256 pKa = 3.4QIAKK260 pKa = 10.16FMLANKK266 pKa = 9.19PAVLTKK272 pKa = 10.45INGYY276 pKa = 10.55LMAIAGYY283 pKa = 8.67TGTRR287 pKa = 11.84EE288 pKa = 4.09QKK290 pKa = 10.54RR291 pKa = 11.84IAFPPGTLAYY301 pKa = 10.34LLHH304 pKa = 7.4DD305 pKa = 4.44LDD307 pKa = 5.5KK308 pKa = 11.32EE309 pKa = 4.4NKK311 pKa = 9.64LHH313 pKa = 6.83FNEE316 pKa = 4.74DD317 pKa = 3.32RR318 pKa = 11.84CSKK321 pKa = 10.52FDD323 pKa = 3.67FSGAGAFRR331 pKa = 11.84MYY333 pKa = 11.15SHH335 pKa = 7.54FLKK338 pKa = 10.59VCKK341 pKa = 10.22KK342 pKa = 10.29NPLRR346 pKa = 11.84MNIADD351 pKa = 4.04PGKK354 pKa = 9.76ARR356 pKa = 11.84QLLFHH361 pKa = 6.46TMFGTHH367 pKa = 5.81VEE369 pKa = 4.28DD370 pKa = 5.25FGILQSMTDD379 pKa = 3.03VSDD382 pKa = 3.09WLKK385 pKa = 11.0RR386 pKa = 11.84KK387 pKa = 10.21DD388 pKa = 3.58FEE390 pKa = 5.14EE391 pKa = 4.1EE392 pKa = 4.1FKK394 pKa = 10.56TLRR397 pKa = 11.84ASHH400 pKa = 6.34ARR402 pKa = 11.84VEE404 pKa = 4.65GNFYY408 pKa = 9.73PIEE411 pKa = 3.67MRR413 pKa = 11.84YY414 pKa = 9.01YY415 pKa = 10.86SKK417 pKa = 11.09VCSSLNTKK425 pKa = 9.66MIGGGSAPVTNCQIFSGNRR444 pKa = 11.84KK445 pKa = 9.74RR446 pKa = 11.84IVTEE450 pKa = 3.95GLKK453 pKa = 10.67SLGQQGVGAGLSALDD468 pKa = 3.72KK469 pKa = 11.37DD470 pKa = 4.73SIEE473 pKa = 4.01KK474 pKa = 10.56LLRR477 pKa = 11.84EE478 pKa = 4.04HH479 pKa = 7.03LEE481 pKa = 4.13SIKK484 pKa = 10.93AAFGDD489 pKa = 4.01SFEE492 pKa = 5.34AGTVKK497 pKa = 9.36WRR499 pKa = 11.84KK500 pKa = 9.21FDD502 pKa = 3.26GLTWDD507 pKa = 3.6KK508 pKa = 11.42EE509 pKa = 4.05GDD511 pKa = 3.85EE512 pKa = 4.49VEE514 pKa = 4.18MRR516 pKa = 11.84PGEE519 pKa = 3.95VGEE522 pKa = 4.92IYY524 pKa = 10.12WSNN527 pKa = 3.04
MM1 pKa = 7.43EE2 pKa = 5.29VDD4 pKa = 4.5PAPSTAPKK12 pKa = 8.41PTRR15 pKa = 11.84KK16 pKa = 9.36RR17 pKa = 11.84VNNSGEE23 pKa = 4.21EE24 pKa = 3.82IGSGGKK30 pKa = 9.41RR31 pKa = 11.84ALTGLSLTQKK41 pKa = 10.2VAIHH45 pKa = 5.83HH46 pKa = 7.14AIALMVYY53 pKa = 9.66HH54 pKa = 5.99VVIDD58 pKa = 3.62IFKK61 pKa = 9.33CTIIDD66 pKa = 5.58LITDD70 pKa = 3.48VNIGVYY76 pKa = 10.71AMNQIWSIHH85 pKa = 5.46NYY87 pKa = 9.26FRR89 pKa = 11.84TKK91 pKa = 10.61NNSGRR96 pKa = 11.84SSIGAKK102 pKa = 9.28IDD104 pKa = 3.55NATDD108 pKa = 3.75TYY110 pKa = 11.41QFILHH115 pKa = 6.34GRR117 pKa = 11.84TITVNKK123 pKa = 10.06EE124 pKa = 3.72EE125 pKa = 4.33LKK127 pKa = 10.8GVYY130 pKa = 8.98KK131 pKa = 10.59RR132 pKa = 11.84CITTYY137 pKa = 10.83FDD139 pKa = 3.42WDD141 pKa = 4.3KK142 pKa = 11.12MNDD145 pKa = 3.09WMGSVHH151 pKa = 7.41TIMALWNLFGSRR163 pKa = 11.84LSEE166 pKa = 3.88VRR168 pKa = 11.84IMPSANTISVEE179 pKa = 3.77KK180 pKa = 10.59DD181 pKa = 2.71NRR183 pKa = 11.84TTVVKK188 pKa = 10.68DD189 pKa = 3.62FNQYY193 pKa = 10.48GIPAGMRR200 pKa = 11.84HH201 pKa = 5.99FATGADD207 pKa = 4.53FKK209 pKa = 10.69PTMKK213 pKa = 10.48SALAQSMGPVTVLVQLSEE231 pKa = 4.6AKK233 pKa = 10.45DD234 pKa = 3.39NQYY237 pKa = 11.0AGKK240 pKa = 9.27WVDD243 pKa = 3.04ALKK246 pKa = 10.54RR247 pKa = 11.84AFSHH251 pKa = 6.57IPHH254 pKa = 7.38IDD256 pKa = 3.4QIAKK260 pKa = 10.16FMLANKK266 pKa = 9.19PAVLTKK272 pKa = 10.45INGYY276 pKa = 10.55LMAIAGYY283 pKa = 8.67TGTRR287 pKa = 11.84EE288 pKa = 4.09QKK290 pKa = 10.54RR291 pKa = 11.84IAFPPGTLAYY301 pKa = 10.34LLHH304 pKa = 7.4DD305 pKa = 4.44LDD307 pKa = 5.5KK308 pKa = 11.32EE309 pKa = 4.4NKK311 pKa = 9.64LHH313 pKa = 6.83FNEE316 pKa = 4.74DD317 pKa = 3.32RR318 pKa = 11.84CSKK321 pKa = 10.52FDD323 pKa = 3.67FSGAGAFRR331 pKa = 11.84MYY333 pKa = 11.15SHH335 pKa = 7.54FLKK338 pKa = 10.59VCKK341 pKa = 10.22KK342 pKa = 10.29NPLRR346 pKa = 11.84MNIADD351 pKa = 4.04PGKK354 pKa = 9.76ARR356 pKa = 11.84QLLFHH361 pKa = 6.46TMFGTHH367 pKa = 5.81VEE369 pKa = 4.28DD370 pKa = 5.25FGILQSMTDD379 pKa = 3.03VSDD382 pKa = 3.09WLKK385 pKa = 11.0RR386 pKa = 11.84KK387 pKa = 10.21DD388 pKa = 3.58FEE390 pKa = 5.14EE391 pKa = 4.1EE392 pKa = 4.1FKK394 pKa = 10.56TLRR397 pKa = 11.84ASHH400 pKa = 6.34ARR402 pKa = 11.84VEE404 pKa = 4.65GNFYY408 pKa = 9.73PIEE411 pKa = 3.67MRR413 pKa = 11.84YY414 pKa = 9.01YY415 pKa = 10.86SKK417 pKa = 11.09VCSSLNTKK425 pKa = 9.66MIGGGSAPVTNCQIFSGNRR444 pKa = 11.84KK445 pKa = 9.74RR446 pKa = 11.84IVTEE450 pKa = 3.95GLKK453 pKa = 10.67SLGQQGVGAGLSALDD468 pKa = 3.72KK469 pKa = 11.37DD470 pKa = 4.73SIEE473 pKa = 4.01KK474 pKa = 10.56LLRR477 pKa = 11.84EE478 pKa = 4.04HH479 pKa = 7.03LEE481 pKa = 4.13SIKK484 pKa = 10.93AAFGDD489 pKa = 4.01SFEE492 pKa = 5.34AGTVKK497 pKa = 9.36WRR499 pKa = 11.84KK500 pKa = 9.21FDD502 pKa = 3.26GLTWDD507 pKa = 3.6KK508 pKa = 11.42EE509 pKa = 4.05GDD511 pKa = 3.85EE512 pKa = 4.49VEE514 pKa = 4.18MRR516 pKa = 11.84PGEE519 pKa = 3.95VGEE522 pKa = 4.92IYY524 pKa = 10.12WSNN527 pKa = 3.04
Molecular weight: 59.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3502 |
266 |
777 |
583.7 |
66.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.368 ± 0.4 | 1.571 ± 0.3 |
5.111 ± 0.29 | 7.453 ± 0.7 |
4.055 ± 0.256 | 6.91 ± 0.565 |
2.513 ± 0.195 | 4.712 ± 0.345 |
6.71 ± 0.308 | 9.338 ± 0.476 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.884 ± 0.207 | 4.397 ± 0.267 |
4.283 ± 0.4 | 3.055 ± 0.157 |
6.139 ± 0.525 | 6.853 ± 0.402 |
5.911 ± 0.169 | 6.425 ± 0.281 |
2.113 ± 0.243 | 3.198 ± 0.357 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
