
Pseudomicrostroma glucosiphilum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Ustilaginomycotina; Exobasidiomycetes; Microstromatales; Microstromatales incertae sedis; Pseudomicrostroma
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
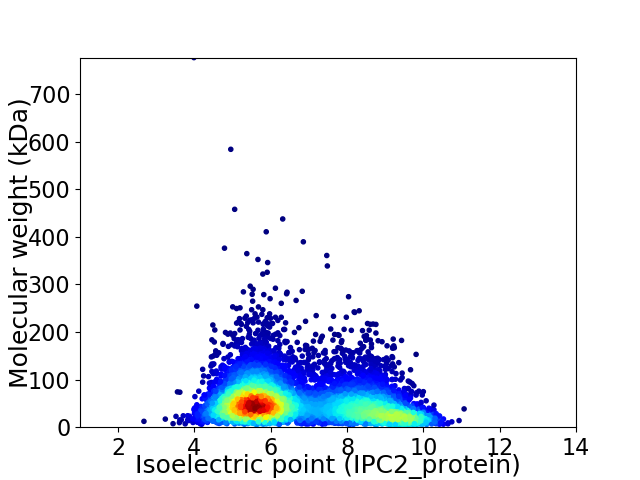
Virtual 2D-PAGE plot for 6679 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316U6N7|A0A316U6N7_9BASI Putative CCT6-component of chaperonin-containing T-complex OS=Pseudomicrostroma glucosiphilum OX=1684307 GN=BCV69DRAFT_283362 PE=3 SV=1
MM1 pKa = 7.64EE2 pKa = 5.3SMINKK7 pKa = 9.39AKK9 pKa = 10.47EE10 pKa = 3.79FASSEE15 pKa = 3.88KK16 pKa = 9.02GQNMMDD22 pKa = 3.14KK23 pKa = 11.0FGGNKK28 pKa = 9.7NNNNNNDD35 pKa = 3.51DD36 pKa = 3.59SSYY39 pKa = 11.61GGNNDD44 pKa = 2.98NSSYY48 pKa = 11.54GGGNDD53 pKa = 3.07NSSYY57 pKa = 11.09GGNDD61 pKa = 3.03NSSGGGGGGFGMQDD75 pKa = 2.98ASSGGDD81 pKa = 3.11SYY83 pKa = 12.02GSGGGQGGFGGDD95 pKa = 3.12TSGQGGYY102 pKa = 9.52GANDD106 pKa = 3.74GGDD109 pKa = 3.91SYY111 pKa = 11.85SSSNQQGGGGYY122 pKa = 9.62GGSGGGQFGDD132 pKa = 4.31DD133 pKa = 3.56SSSGGGRR140 pKa = 11.84QGGGGFGGGGQDD152 pKa = 3.97DD153 pKa = 4.69FSSGGARR160 pKa = 11.84QGGGGGFGGGGQDD173 pKa = 4.18DD174 pKa = 4.49FSSGGGQGGQGGGFGGDD191 pKa = 3.04TSGQGGYY198 pKa = 9.52GANDD202 pKa = 3.79GGDD205 pKa = 3.68SFSSSNQQGGGYY217 pKa = 9.19GGSGGGQFGDD227 pKa = 4.15DD228 pKa = 3.83SGSGGGGGGYY238 pKa = 10.48GGGGRR243 pKa = 11.84DD244 pKa = 3.43DD245 pKa = 3.93QYY247 pKa = 12.23
MM1 pKa = 7.64EE2 pKa = 5.3SMINKK7 pKa = 9.39AKK9 pKa = 10.47EE10 pKa = 3.79FASSEE15 pKa = 3.88KK16 pKa = 9.02GQNMMDD22 pKa = 3.14KK23 pKa = 11.0FGGNKK28 pKa = 9.7NNNNNNDD35 pKa = 3.51DD36 pKa = 3.59SSYY39 pKa = 11.61GGNNDD44 pKa = 2.98NSSYY48 pKa = 11.54GGGNDD53 pKa = 3.07NSSYY57 pKa = 11.09GGNDD61 pKa = 3.03NSSGGGGGGFGMQDD75 pKa = 2.98ASSGGDD81 pKa = 3.11SYY83 pKa = 12.02GSGGGQGGFGGDD95 pKa = 3.12TSGQGGYY102 pKa = 9.52GANDD106 pKa = 3.74GGDD109 pKa = 3.91SYY111 pKa = 11.85SSSNQQGGGGYY122 pKa = 9.62GGSGGGQFGDD132 pKa = 4.31DD133 pKa = 3.56SSSGGGRR140 pKa = 11.84QGGGGFGGGGQDD152 pKa = 3.97DD153 pKa = 4.69FSSGGARR160 pKa = 11.84QGGGGGFGGGGQDD173 pKa = 4.18DD174 pKa = 4.49FSSGGGQGGQGGGFGGDD191 pKa = 3.04TSGQGGYY198 pKa = 9.52GANDD202 pKa = 3.79GGDD205 pKa = 3.68SFSSSNQQGGGYY217 pKa = 9.19GGSGGGQFGDD227 pKa = 4.15DD228 pKa = 3.83SGSGGGGGGYY238 pKa = 10.48GGGGRR243 pKa = 11.84DD244 pKa = 3.43DD245 pKa = 3.93QYY247 pKa = 12.23
Molecular weight: 22.72 kDa
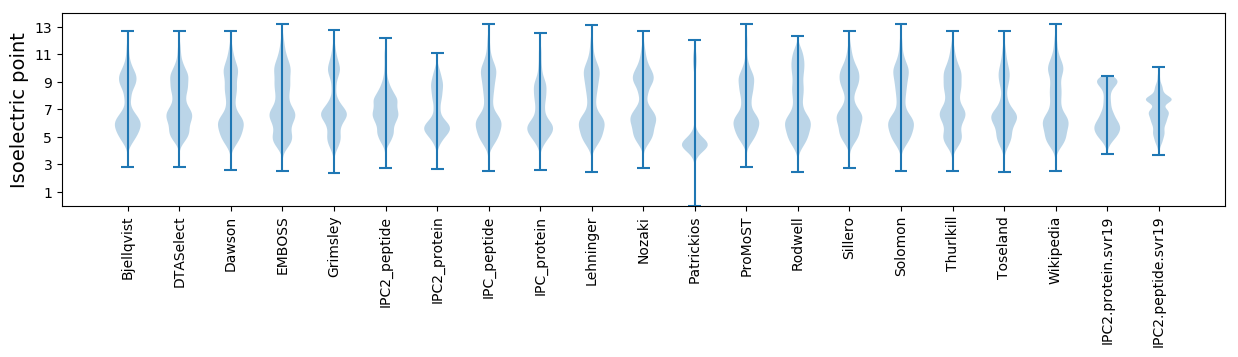
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316U8R9|A0A316U8R9_9BASI Threonine synthase OS=Pseudomicrostroma glucosiphilum OX=1684307 GN=BCV69DRAFT_282335 PE=3 SV=1
MM1 pKa = 7.97DD2 pKa = 5.0GALAAAIKK10 pKa = 9.89QGKK13 pKa = 8.37ALKK16 pKa = 10.02KK17 pKa = 10.24AQTNDD22 pKa = 2.75RR23 pKa = 11.84SAPAIAGSVGGSGGGGRR40 pKa = 11.84PAGSAPPVPGAAAASAMASAAVTKK64 pKa = 7.32MTSSGPPNLGGLFAGGMPTLRR85 pKa = 11.84SASGGSTAPKK95 pKa = 9.84PPGRR99 pKa = 11.84PAPPPASSAPAQPPPARR116 pKa = 11.84GVPTPPRR123 pKa = 11.84ASAPPPPAPPNRR135 pKa = 11.84PTAAAPPPPPGRR147 pKa = 11.84PAPPASGPPPPPPPPPGRR165 pKa = 11.84PASSAPPPPTRR176 pKa = 11.84PTSSAPPPPPPPPAAPPTPSRR197 pKa = 11.84STLAPAAPPPPPPPPGPSKK216 pKa = 10.64GAPPPPSRR224 pKa = 11.84PSAVPPRR231 pKa = 11.84APPPPQPQTSAPSTPSRR248 pKa = 11.84SVPAPPSRR256 pKa = 11.84PAPGASAAPPPAPPPPPPPSAAPSTPSRR284 pKa = 11.84SVPTAPSRR292 pKa = 11.84LAPSGASAAPPPPAPSRR309 pKa = 11.84LAPSGASAAPPPPAPPPPPPASRR332 pKa = 11.84APAPPPPSRR341 pKa = 11.84APAPPPPSRR350 pKa = 11.84STPTSRR356 pKa = 11.84PPPPPVAASAATNGSGRR373 pKa = 11.84STPITNGANGAAPPLRR389 pKa = 11.84NSIVGPPSNGYY400 pKa = 9.03GAGGGGGGGRR410 pKa = 11.84SKK412 pKa = 11.34
MM1 pKa = 7.97DD2 pKa = 5.0GALAAAIKK10 pKa = 9.89QGKK13 pKa = 8.37ALKK16 pKa = 10.02KK17 pKa = 10.24AQTNDD22 pKa = 2.75RR23 pKa = 11.84SAPAIAGSVGGSGGGGRR40 pKa = 11.84PAGSAPPVPGAAAASAMASAAVTKK64 pKa = 7.32MTSSGPPNLGGLFAGGMPTLRR85 pKa = 11.84SASGGSTAPKK95 pKa = 9.84PPGRR99 pKa = 11.84PAPPPASSAPAQPPPARR116 pKa = 11.84GVPTPPRR123 pKa = 11.84ASAPPPPAPPNRR135 pKa = 11.84PTAAAPPPPPGRR147 pKa = 11.84PAPPASGPPPPPPPPPGRR165 pKa = 11.84PASSAPPPPTRR176 pKa = 11.84PTSSAPPPPPPPPAAPPTPSRR197 pKa = 11.84STLAPAAPPPPPPPPGPSKK216 pKa = 10.64GAPPPPSRR224 pKa = 11.84PSAVPPRR231 pKa = 11.84APPPPQPQTSAPSTPSRR248 pKa = 11.84SVPAPPSRR256 pKa = 11.84PAPGASAAPPPAPPPPPPPSAAPSTPSRR284 pKa = 11.84SVPTAPSRR292 pKa = 11.84LAPSGASAAPPPPAPSRR309 pKa = 11.84LAPSGASAAPPPPAPPPPPPASRR332 pKa = 11.84APAPPPPSRR341 pKa = 11.84APAPPPPSRR350 pKa = 11.84STPTSRR356 pKa = 11.84PPPPPVAASAATNGSGRR373 pKa = 11.84STPITNGANGAAPPLRR389 pKa = 11.84NSIVGPPSNGYY400 pKa = 9.03GAGGGGGGGRR410 pKa = 11.84SKK412 pKa = 11.34
Molecular weight: 38.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3653858 |
50 |
9118 |
547.1 |
59.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.411 ± 0.095 | 0.992 ± 0.011 |
5.343 ± 0.021 | 6.259 ± 0.031 |
3.033 ± 0.017 | 7.855 ± 0.031 |
2.241 ± 0.014 | 3.804 ± 0.02 |
4.431 ± 0.027 | 8.928 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.866 ± 0.01 | 2.805 ± 0.013 |
6.227 ± 0.032 | 4.114 ± 0.023 |
6.587 ± 0.03 | 10.199 ± 0.075 |
5.775 ± 0.026 | 5.693 ± 0.02 |
1.238 ± 0.009 | 2.196 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
