
Chengkuizengella sp. YPA3-1-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Chengkuizengella
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
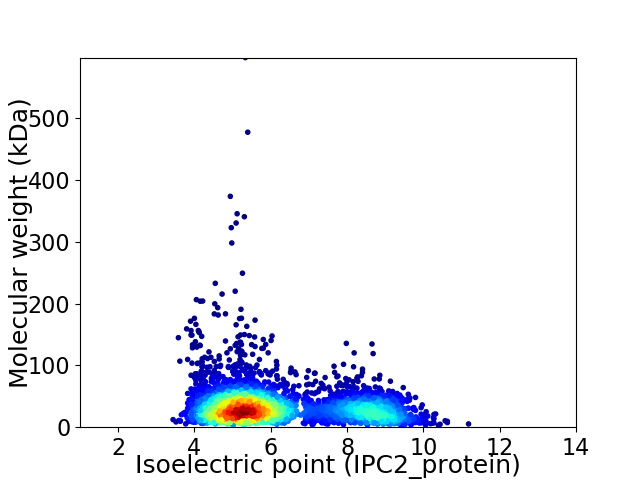
Virtual 2D-PAGE plot for 3947 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N9Q360|A0A6N9Q360_9BACL SLC13 family permease OS=Chengkuizengella sp. YPA3-1-1 OX=2507566 GN=ERL59_09725 PE=4 SV=1
MM1 pKa = 8.31DD2 pKa = 6.26DD3 pKa = 3.53FNEE6 pKa = 4.67WIQWNANQNDD16 pKa = 4.0LEE18 pKa = 4.19QAIVRR23 pKa = 11.84NGGTYY28 pKa = 10.26NLEE31 pKa = 4.43GNTNLYY37 pKa = 10.86FFFNGGNTPEE47 pKa = 5.21SFDD50 pKa = 3.36QFINRR55 pKa = 11.84DD56 pKa = 2.95ISDD59 pKa = 4.02YY60 pKa = 10.53DD61 pKa = 3.66TLEE64 pKa = 4.17LVLKK68 pKa = 9.92GGSGGEE74 pKa = 3.67EE75 pKa = 3.79QYY77 pKa = 11.5INLVLNDD84 pKa = 4.63DD85 pKa = 4.55SNHH88 pKa = 5.87SLSLSNYY95 pKa = 9.32GNLTANYY102 pKa = 8.45STIQIPLADD111 pKa = 4.64FNANLTNVKK120 pKa = 9.99FIRR123 pKa = 11.84FEE125 pKa = 4.19GVGTAKK131 pKa = 10.12IVRR134 pKa = 11.84IDD136 pKa = 4.45DD137 pKa = 4.29MILKK141 pKa = 8.28YY142 pKa = 9.48TGISPTDD149 pKa = 3.52PTDD152 pKa = 3.59PTDD155 pKa = 3.84PTDD158 pKa = 3.84PTDD161 pKa = 3.84PTDD164 pKa = 3.77PTDD167 pKa = 3.62PTEE170 pKa = 4.48PTDD173 pKa = 3.61PTDD176 pKa = 4.2PGDD179 pKa = 3.74VPDD182 pKa = 5.09FPVGSGVMTFQLQNGTNGQYY202 pKa = 10.66SDD204 pKa = 5.6DD205 pKa = 4.14EE206 pKa = 4.86IYY208 pKa = 9.79WAIYY212 pKa = 9.62GYY214 pKa = 10.82NKK216 pKa = 8.71EE217 pKa = 4.4TGQLSYY223 pKa = 11.68VDD225 pKa = 4.32ANGNLIASSVSDD237 pKa = 3.5NDD239 pKa = 4.13APGHH243 pKa = 4.79LTKK246 pKa = 10.98NGEE249 pKa = 3.85NYY251 pKa = 10.87ANYY254 pKa = 8.71FNKK257 pKa = 9.65MSEE260 pKa = 4.29TDD262 pKa = 3.37WVSMPEE268 pKa = 3.8IDD270 pKa = 3.83SGRR273 pKa = 11.84MFISMGSPMYY283 pKa = 10.41IKK285 pKa = 10.82LNIAADD291 pKa = 3.79GRR293 pKa = 11.84VGFAGPDD300 pKa = 3.57LNNPTDD306 pKa = 4.35PNQDD310 pKa = 2.95IYY312 pKa = 11.27FEE314 pKa = 4.68WIEE317 pKa = 3.87FTIDD321 pKa = 2.47QWGYY325 pKa = 9.66HH326 pKa = 5.54GNTTRR331 pKa = 11.84VDD333 pKa = 3.42QFSFPITTRR342 pKa = 11.84LVASDD347 pKa = 4.02GYY349 pKa = 11.26DD350 pKa = 3.15RR351 pKa = 11.84IVGEE355 pKa = 4.61TYY357 pKa = 10.18TRR359 pKa = 11.84DD360 pKa = 3.48QIFTAFKK367 pKa = 11.04NEE369 pKa = 4.04MPTEE373 pKa = 4.03FQTLVEE379 pKa = 4.09EE380 pKa = 5.25PYY382 pKa = 10.63RR383 pKa = 11.84IVAPGKK389 pKa = 10.99GEE391 pKa = 4.11FKK393 pKa = 10.88PEE395 pKa = 4.16GIYY398 pKa = 10.98ANYY401 pKa = 9.33FDD403 pKa = 5.43NYY405 pKa = 9.92VNEE408 pKa = 3.76VWDD411 pKa = 4.9FYY413 pKa = 10.83RR414 pKa = 11.84TNEE417 pKa = 3.9LTFTAEE423 pKa = 3.71AGTFSGHH430 pKa = 5.19VVGDD434 pKa = 3.54DD435 pKa = 3.78FVFSKK440 pKa = 11.18NGGPYY445 pKa = 10.72DD446 pKa = 3.51LFIRR450 pKa = 11.84NKK452 pKa = 8.64PTTLQVFEE460 pKa = 5.34CSGPMDD466 pKa = 3.84TGSPDD471 pKa = 3.43EE472 pKa = 4.71KK473 pKa = 10.87VVEE476 pKa = 4.34AQVCAALNRR485 pKa = 11.84GMMYY489 pKa = 10.78DD490 pKa = 3.56PANWGNSDD498 pKa = 4.75FFYY501 pKa = 11.18QNNTANFYY509 pKa = 11.17AKK511 pKa = 10.03FWHH514 pKa = 7.41DD515 pKa = 3.48YY516 pKa = 10.93SIDD519 pKa = 3.24GLAYY523 pKa = 9.77GFAYY527 pKa = 10.48DD528 pKa = 4.64DD529 pKa = 3.55VRR531 pKa = 11.84NYY533 pKa = 9.29STLLEE538 pKa = 4.21HH539 pKa = 7.37HH540 pKa = 7.36DD541 pKa = 4.04PDD543 pKa = 3.84VLFINVGWW551 pKa = 3.67
MM1 pKa = 8.31DD2 pKa = 6.26DD3 pKa = 3.53FNEE6 pKa = 4.67WIQWNANQNDD16 pKa = 4.0LEE18 pKa = 4.19QAIVRR23 pKa = 11.84NGGTYY28 pKa = 10.26NLEE31 pKa = 4.43GNTNLYY37 pKa = 10.86FFFNGGNTPEE47 pKa = 5.21SFDD50 pKa = 3.36QFINRR55 pKa = 11.84DD56 pKa = 2.95ISDD59 pKa = 4.02YY60 pKa = 10.53DD61 pKa = 3.66TLEE64 pKa = 4.17LVLKK68 pKa = 9.92GGSGGEE74 pKa = 3.67EE75 pKa = 3.79QYY77 pKa = 11.5INLVLNDD84 pKa = 4.63DD85 pKa = 4.55SNHH88 pKa = 5.87SLSLSNYY95 pKa = 9.32GNLTANYY102 pKa = 8.45STIQIPLADD111 pKa = 4.64FNANLTNVKK120 pKa = 9.99FIRR123 pKa = 11.84FEE125 pKa = 4.19GVGTAKK131 pKa = 10.12IVRR134 pKa = 11.84IDD136 pKa = 4.45DD137 pKa = 4.29MILKK141 pKa = 8.28YY142 pKa = 9.48TGISPTDD149 pKa = 3.52PTDD152 pKa = 3.59PTDD155 pKa = 3.84PTDD158 pKa = 3.84PTDD161 pKa = 3.84PTDD164 pKa = 3.77PTDD167 pKa = 3.62PTEE170 pKa = 4.48PTDD173 pKa = 3.61PTDD176 pKa = 4.2PGDD179 pKa = 3.74VPDD182 pKa = 5.09FPVGSGVMTFQLQNGTNGQYY202 pKa = 10.66SDD204 pKa = 5.6DD205 pKa = 4.14EE206 pKa = 4.86IYY208 pKa = 9.79WAIYY212 pKa = 9.62GYY214 pKa = 10.82NKK216 pKa = 8.71EE217 pKa = 4.4TGQLSYY223 pKa = 11.68VDD225 pKa = 4.32ANGNLIASSVSDD237 pKa = 3.5NDD239 pKa = 4.13APGHH243 pKa = 4.79LTKK246 pKa = 10.98NGEE249 pKa = 3.85NYY251 pKa = 10.87ANYY254 pKa = 8.71FNKK257 pKa = 9.65MSEE260 pKa = 4.29TDD262 pKa = 3.37WVSMPEE268 pKa = 3.8IDD270 pKa = 3.83SGRR273 pKa = 11.84MFISMGSPMYY283 pKa = 10.41IKK285 pKa = 10.82LNIAADD291 pKa = 3.79GRR293 pKa = 11.84VGFAGPDD300 pKa = 3.57LNNPTDD306 pKa = 4.35PNQDD310 pKa = 2.95IYY312 pKa = 11.27FEE314 pKa = 4.68WIEE317 pKa = 3.87FTIDD321 pKa = 2.47QWGYY325 pKa = 9.66HH326 pKa = 5.54GNTTRR331 pKa = 11.84VDD333 pKa = 3.42QFSFPITTRR342 pKa = 11.84LVASDD347 pKa = 4.02GYY349 pKa = 11.26DD350 pKa = 3.15RR351 pKa = 11.84IVGEE355 pKa = 4.61TYY357 pKa = 10.18TRR359 pKa = 11.84DD360 pKa = 3.48QIFTAFKK367 pKa = 11.04NEE369 pKa = 4.04MPTEE373 pKa = 4.03FQTLVEE379 pKa = 4.09EE380 pKa = 5.25PYY382 pKa = 10.63RR383 pKa = 11.84IVAPGKK389 pKa = 10.99GEE391 pKa = 4.11FKK393 pKa = 10.88PEE395 pKa = 4.16GIYY398 pKa = 10.98ANYY401 pKa = 9.33FDD403 pKa = 5.43NYY405 pKa = 9.92VNEE408 pKa = 3.76VWDD411 pKa = 4.9FYY413 pKa = 10.83RR414 pKa = 11.84TNEE417 pKa = 3.9LTFTAEE423 pKa = 3.71AGTFSGHH430 pKa = 5.19VVGDD434 pKa = 3.54DD435 pKa = 3.78FVFSKK440 pKa = 11.18NGGPYY445 pKa = 10.72DD446 pKa = 3.51LFIRR450 pKa = 11.84NKK452 pKa = 8.64PTTLQVFEE460 pKa = 5.34CSGPMDD466 pKa = 3.84TGSPDD471 pKa = 3.43EE472 pKa = 4.71KK473 pKa = 10.87VVEE476 pKa = 4.34AQVCAALNRR485 pKa = 11.84GMMYY489 pKa = 10.78DD490 pKa = 3.56PANWGNSDD498 pKa = 4.75FFYY501 pKa = 11.18QNNTANFYY509 pKa = 11.17AKK511 pKa = 10.03FWHH514 pKa = 7.41DD515 pKa = 3.48YY516 pKa = 10.93SIDD519 pKa = 3.24GLAYY523 pKa = 9.77GFAYY527 pKa = 10.48DD528 pKa = 4.64DD529 pKa = 3.55VRR531 pKa = 11.84NYY533 pKa = 9.29STLLEE538 pKa = 4.21HH539 pKa = 7.37HH540 pKa = 7.36DD541 pKa = 4.04PDD543 pKa = 3.84VLFINVGWW551 pKa = 3.67
Molecular weight: 62.02 kDa
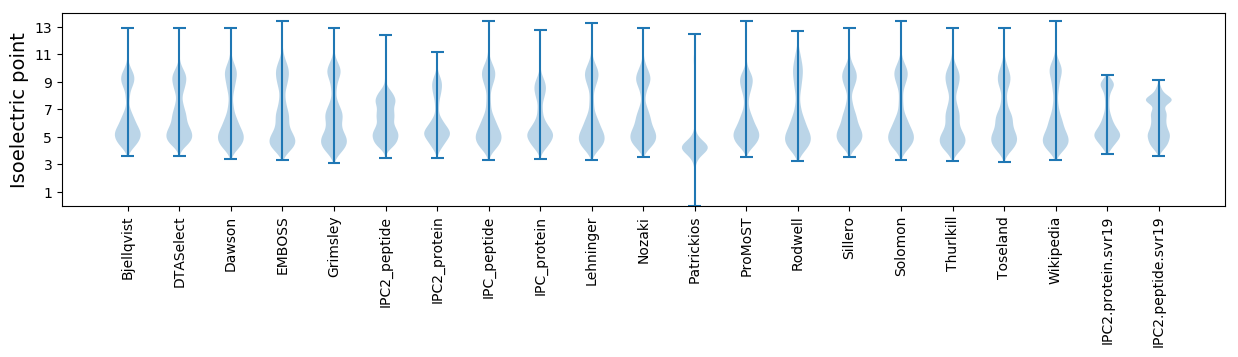
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N9Q307|A0A6N9Q307_9BACL Membrane protein insertase YidC OS=Chengkuizengella sp. YPA3-1-1 OX=2507566 GN=yidC PE=3 SV=1
MM1 pKa = 7.6KK2 pKa = 8.72PTFQPNTRR10 pKa = 11.84KK11 pKa = 9.78RR12 pKa = 11.84KK13 pKa = 8.71KK14 pKa = 8.74VHH16 pKa = 5.67GFRR19 pKa = 11.84KK20 pKa = 9.98RR21 pKa = 11.84MNSKK25 pKa = 10.17NGRR28 pKa = 11.84KK29 pKa = 9.32VLTARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.12GRR39 pKa = 11.84KK40 pKa = 8.67ILSAA44 pKa = 4.02
MM1 pKa = 7.6KK2 pKa = 8.72PTFQPNTRR10 pKa = 11.84KK11 pKa = 9.78RR12 pKa = 11.84KK13 pKa = 8.71KK14 pKa = 8.74VHH16 pKa = 5.67GFRR19 pKa = 11.84KK20 pKa = 9.98RR21 pKa = 11.84MNSKK25 pKa = 10.17NGRR28 pKa = 11.84KK29 pKa = 9.32VLTARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.12GRR39 pKa = 11.84KK40 pKa = 8.67ILSAA44 pKa = 4.02
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1225611 |
18 |
5234 |
310.5 |
35.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.582 ± 0.042 | 0.773 ± 0.016 |
5.466 ± 0.036 | 7.522 ± 0.048 |
4.548 ± 0.036 | 6.362 ± 0.038 |
2.072 ± 0.02 | 8.69 ± 0.041 |
7.157 ± 0.044 | 9.467 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.64 ± 0.018 | 5.45 ± 0.04 |
3.388 ± 0.021 | 3.877 ± 0.029 |
3.533 ± 0.027 | 6.485 ± 0.034 |
5.437 ± 0.036 | 6.653 ± 0.031 |
1.071 ± 0.016 | 3.828 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
