
Capybara microvirus Cap3_SP_469
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.28
Get precalculated fractions of proteins
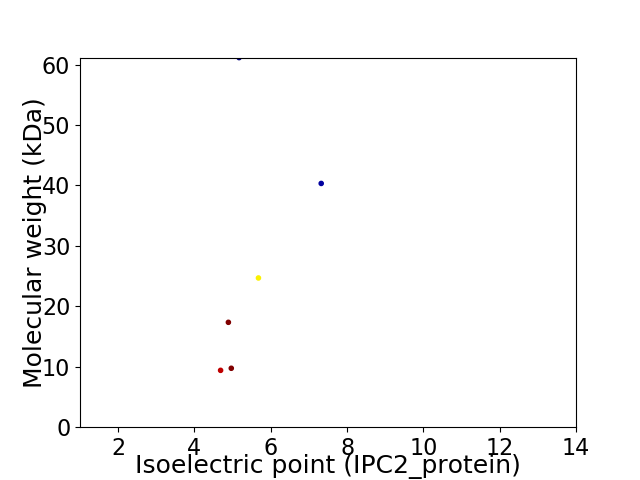
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8P9|A0A4P8W8P9_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_469 OX=2585464 PE=4 SV=1
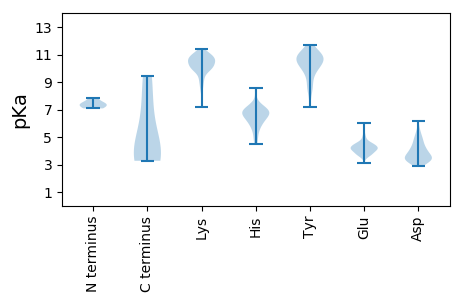
MM1 pKa = 7.37SKK3 pKa = 10.61KK4 pKa = 10.41KK5 pKa = 10.42VVNDD9 pKa = 3.64LSYY12 pKa = 10.3HH13 pKa = 4.53YY14 pKa = 10.38HH15 pKa = 6.64VNTCDD20 pKa = 3.16EE21 pKa = 5.08GINEE25 pKa = 4.24VKK27 pKa = 10.43CACKK31 pKa = 10.21LYY33 pKa = 11.14NEE35 pKa = 4.53GKK37 pKa = 8.5YY38 pKa = 8.39TFEE41 pKa = 4.05EE42 pKa = 5.06LILFLNTIKK51 pKa = 9.37TQIDD55 pKa = 3.98DD56 pKa = 4.86DD57 pKa = 5.23LDD59 pKa = 4.15DD60 pKa = 6.18LILEE64 pKa = 4.52VICDD68 pKa = 4.19RR69 pKa = 11.84IEE71 pKa = 4.38KK72 pKa = 10.17IEE74 pKa = 4.25SEE76 pKa = 4.65VYY78 pKa = 10.73KK79 pKa = 11.1NN80 pKa = 3.27
MM1 pKa = 7.37SKK3 pKa = 10.61KK4 pKa = 10.41KK5 pKa = 10.42VVNDD9 pKa = 3.64LSYY12 pKa = 10.3HH13 pKa = 4.53YY14 pKa = 10.38HH15 pKa = 6.64VNTCDD20 pKa = 3.16EE21 pKa = 5.08GINEE25 pKa = 4.24VKK27 pKa = 10.43CACKK31 pKa = 10.21LYY33 pKa = 11.14NEE35 pKa = 4.53GKK37 pKa = 8.5YY38 pKa = 8.39TFEE41 pKa = 4.05EE42 pKa = 5.06LILFLNTIKK51 pKa = 9.37TQIDD55 pKa = 3.98DD56 pKa = 4.86DD57 pKa = 5.23LDD59 pKa = 4.15DD60 pKa = 6.18LILEE64 pKa = 4.52VICDD68 pKa = 4.19RR69 pKa = 11.84IEE71 pKa = 4.38KK72 pKa = 10.17IEE74 pKa = 4.25SEE76 pKa = 4.65VYY78 pKa = 10.73KK79 pKa = 11.1NN80 pKa = 3.27
Molecular weight: 9.41 kDa
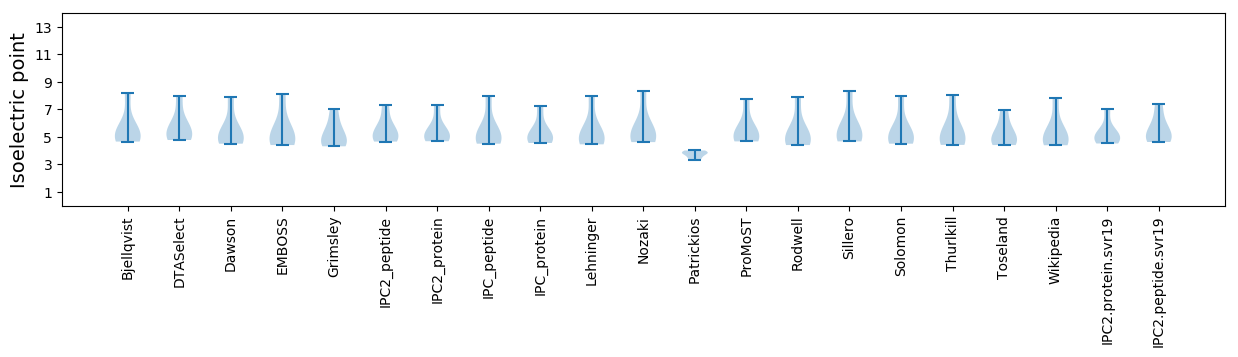
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W549|A0A4P8W549_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_469 OX=2585464 PE=3 SV=1
MM1 pKa = 7.14CTSPLKK7 pKa = 10.52GFPYY11 pKa = 10.83GKK13 pKa = 8.58TKK15 pKa = 10.58NGKK18 pKa = 9.25DD19 pKa = 3.17NFVITSYY26 pKa = 10.89KK27 pKa = 9.68CDD29 pKa = 3.95HH30 pKa = 6.83IEE32 pKa = 3.92VNRR35 pKa = 11.84NDD37 pKa = 3.81VPIKK41 pKa = 10.1IYY43 pKa = 10.58EE44 pKa = 3.92PTFIHH49 pKa = 6.62SQDD52 pKa = 3.16TKK54 pKa = 11.02EE55 pKa = 4.24IIRR58 pKa = 11.84DD59 pKa = 4.41FIEE62 pKa = 4.66IPCGHH67 pKa = 6.86CLEE70 pKa = 4.91CRR72 pKa = 11.84LDD74 pKa = 4.59YY75 pKa = 10.96SRR77 pKa = 11.84QWANRR82 pKa = 11.84CMLEE86 pKa = 4.07AQSYY90 pKa = 7.16KK91 pKa = 10.52HH92 pKa = 6.32NYY94 pKa = 8.68FVTLTYY100 pKa = 11.11DD101 pKa = 4.05DD102 pKa = 4.4LHH104 pKa = 8.53LPMNEE109 pKa = 3.96YY110 pKa = 10.92VDD112 pKa = 3.7VDD114 pKa = 3.77GVLQLSPTLVKK125 pKa = 10.28RR126 pKa = 11.84DD127 pKa = 3.43LQLFMKK133 pKa = 10.54RR134 pKa = 11.84LRR136 pKa = 11.84KK137 pKa = 9.97KK138 pKa = 10.27FGDD141 pKa = 3.44NIRR144 pKa = 11.84FYY146 pKa = 11.58ACGEE150 pKa = 4.29YY151 pKa = 10.59GSKK154 pKa = 7.16THH156 pKa = 6.81RR157 pKa = 11.84PHH159 pKa = 5.64YY160 pKa = 10.36HH161 pKa = 6.98LILFNFNPNDD171 pKa = 5.11LIFLKK176 pKa = 10.91QNFQGDD182 pKa = 4.44LYY184 pKa = 8.88YY185 pKa = 10.47TSPGLEE191 pKa = 4.18SCWKK195 pKa = 9.83YY196 pKa = 10.38GYY198 pKa = 11.62SMIAEE203 pKa = 4.53VNWNTCAYY211 pKa = 7.52VARR214 pKa = 11.84YY215 pKa = 8.62IVKK218 pKa = 10.03KK219 pKa = 10.64QFGEE223 pKa = 3.81NADD226 pKa = 3.63YY227 pKa = 11.25KK228 pKa = 10.66KK229 pKa = 10.87FNYY232 pKa = 9.77EE233 pKa = 3.81PEE235 pKa = 4.12FTLMSRR241 pKa = 11.84KK242 pKa = 9.57PGIARR247 pKa = 11.84DD248 pKa = 3.23WYY250 pKa = 10.29EE251 pKa = 3.63NHH253 pKa = 7.3KK254 pKa = 11.15DD255 pKa = 3.39NLYY258 pKa = 10.73DD259 pKa = 3.6YY260 pKa = 10.79DD261 pKa = 5.68AIYY264 pKa = 10.32IKK266 pKa = 10.67DD267 pKa = 4.61GIPVKK272 pKa = 9.93PSKK275 pKa = 11.1YY276 pKa = 9.74FDD278 pKa = 3.35RR279 pKa = 11.84LYY281 pKa = 10.86DD282 pKa = 3.38IDD284 pKa = 4.94YY285 pKa = 9.04SAQMEE290 pKa = 4.61EE291 pKa = 3.87IKK293 pKa = 10.52KK294 pKa = 10.0VRR296 pKa = 11.84LEE298 pKa = 4.1SMLHH302 pKa = 5.36HH303 pKa = 6.89KK304 pKa = 9.78MLIAEE309 pKa = 4.54MTSQTYY315 pKa = 9.9EE316 pKa = 4.05EE317 pKa = 4.23QLSTRR322 pKa = 11.84NNVIEE327 pKa = 4.46EE328 pKa = 4.14KK329 pKa = 9.98TKK331 pKa = 9.84KK332 pKa = 10.13LKK334 pKa = 10.55RR335 pKa = 11.84KK336 pKa = 9.45EE337 pKa = 3.68II338 pKa = 3.99
MM1 pKa = 7.14CTSPLKK7 pKa = 10.52GFPYY11 pKa = 10.83GKK13 pKa = 8.58TKK15 pKa = 10.58NGKK18 pKa = 9.25DD19 pKa = 3.17NFVITSYY26 pKa = 10.89KK27 pKa = 9.68CDD29 pKa = 3.95HH30 pKa = 6.83IEE32 pKa = 3.92VNRR35 pKa = 11.84NDD37 pKa = 3.81VPIKK41 pKa = 10.1IYY43 pKa = 10.58EE44 pKa = 3.92PTFIHH49 pKa = 6.62SQDD52 pKa = 3.16TKK54 pKa = 11.02EE55 pKa = 4.24IIRR58 pKa = 11.84DD59 pKa = 4.41FIEE62 pKa = 4.66IPCGHH67 pKa = 6.86CLEE70 pKa = 4.91CRR72 pKa = 11.84LDD74 pKa = 4.59YY75 pKa = 10.96SRR77 pKa = 11.84QWANRR82 pKa = 11.84CMLEE86 pKa = 4.07AQSYY90 pKa = 7.16KK91 pKa = 10.52HH92 pKa = 6.32NYY94 pKa = 8.68FVTLTYY100 pKa = 11.11DD101 pKa = 4.05DD102 pKa = 4.4LHH104 pKa = 8.53LPMNEE109 pKa = 3.96YY110 pKa = 10.92VDD112 pKa = 3.7VDD114 pKa = 3.77GVLQLSPTLVKK125 pKa = 10.28RR126 pKa = 11.84DD127 pKa = 3.43LQLFMKK133 pKa = 10.54RR134 pKa = 11.84LRR136 pKa = 11.84KK137 pKa = 9.97KK138 pKa = 10.27FGDD141 pKa = 3.44NIRR144 pKa = 11.84FYY146 pKa = 11.58ACGEE150 pKa = 4.29YY151 pKa = 10.59GSKK154 pKa = 7.16THH156 pKa = 6.81RR157 pKa = 11.84PHH159 pKa = 5.64YY160 pKa = 10.36HH161 pKa = 6.98LILFNFNPNDD171 pKa = 5.11LIFLKK176 pKa = 10.91QNFQGDD182 pKa = 4.44LYY184 pKa = 8.88YY185 pKa = 10.47TSPGLEE191 pKa = 4.18SCWKK195 pKa = 9.83YY196 pKa = 10.38GYY198 pKa = 11.62SMIAEE203 pKa = 4.53VNWNTCAYY211 pKa = 7.52VARR214 pKa = 11.84YY215 pKa = 8.62IVKK218 pKa = 10.03KK219 pKa = 10.64QFGEE223 pKa = 3.81NADD226 pKa = 3.63YY227 pKa = 11.25KK228 pKa = 10.66KK229 pKa = 10.87FNYY232 pKa = 9.77EE233 pKa = 3.81PEE235 pKa = 4.12FTLMSRR241 pKa = 11.84KK242 pKa = 9.57PGIARR247 pKa = 11.84DD248 pKa = 3.23WYY250 pKa = 10.29EE251 pKa = 3.63NHH253 pKa = 7.3KK254 pKa = 11.15DD255 pKa = 3.39NLYY258 pKa = 10.73DD259 pKa = 3.6YY260 pKa = 10.79DD261 pKa = 5.68AIYY264 pKa = 10.32IKK266 pKa = 10.67DD267 pKa = 4.61GIPVKK272 pKa = 9.93PSKK275 pKa = 11.1YY276 pKa = 9.74FDD278 pKa = 3.35RR279 pKa = 11.84LYY281 pKa = 10.86DD282 pKa = 3.38IDD284 pKa = 4.94YY285 pKa = 9.04SAQMEE290 pKa = 4.61EE291 pKa = 3.87IKK293 pKa = 10.52KK294 pKa = 10.0VRR296 pKa = 11.84LEE298 pKa = 4.1SMLHH302 pKa = 5.36HH303 pKa = 6.89KK304 pKa = 9.78MLIAEE309 pKa = 4.54MTSQTYY315 pKa = 9.9EE316 pKa = 4.05EE317 pKa = 4.23QLSTRR322 pKa = 11.84NNVIEE327 pKa = 4.46EE328 pKa = 4.14KK329 pKa = 9.98TKK331 pKa = 9.84KK332 pKa = 10.13LKK334 pKa = 10.55RR335 pKa = 11.84KK336 pKa = 9.45EE337 pKa = 3.68II338 pKa = 3.99
Molecular weight: 40.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1444 |
80 |
552 |
240.7 |
27.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.055 ± 1.099 | 1.316 ± 0.584 |
5.886 ± 0.785 | 5.748 ± 1.157 |
4.571 ± 0.572 | 7.064 ± 1.283 |
1.87 ± 0.487 | 5.886 ± 0.87 |
5.956 ± 1.637 | 7.964 ± 0.45 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.77 ± 0.6 | 8.102 ± 1.357 |
4.155 ± 0.802 | 3.463 ± 0.393 |
3.67 ± 0.709 | 8.657 ± 1.706 |
5.471 ± 0.264 | 5.956 ± 0.6 |
1.108 ± 0.248 | 5.332 ± 0.784 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
