
Salegentibacter echinorum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Salegentibacter
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
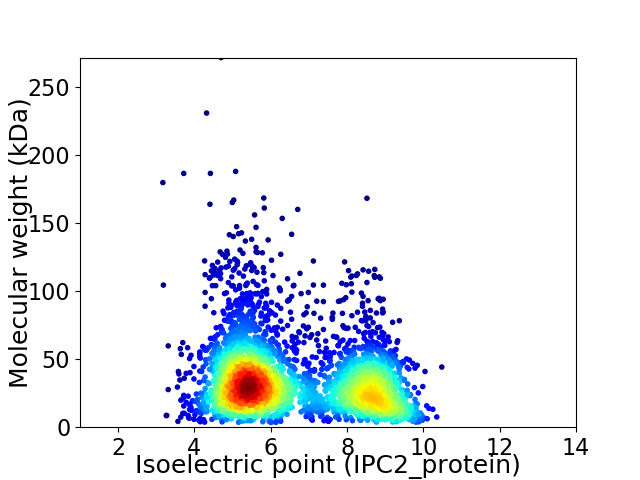
Virtual 2D-PAGE plot for 3479 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
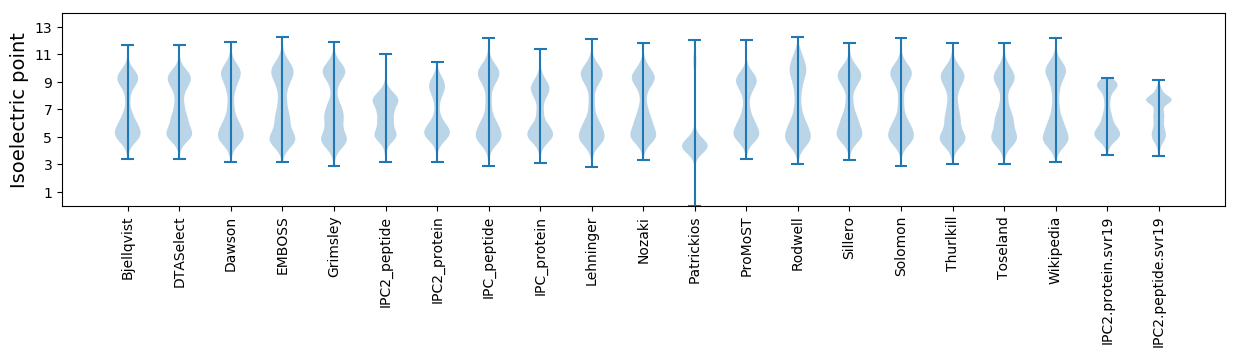
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5FDQ3|A0A1M5FDQ3_9FLAO Acyl carrier protein phosphodiesterase OS=Salegentibacter echinorum OX=1073325 GN=SAMN05444483_103148 PE=4 SV=1
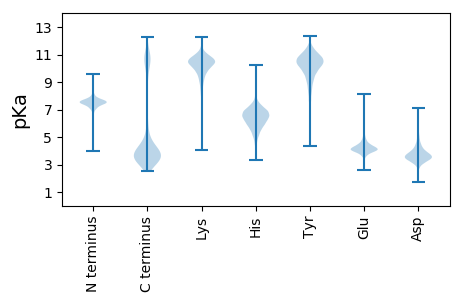
MM1 pKa = 7.35NKK3 pKa = 9.44YY4 pKa = 10.05HH5 pKa = 6.93SLPTGPNFRR14 pKa = 11.84YY15 pKa = 9.57LPKK18 pKa = 10.57GLYY21 pKa = 8.97PLLFIFFFLLKK32 pKa = 10.29SGVGFGQEE40 pKa = 3.65NCIITSTATDD50 pKa = 3.7PTEE53 pKa = 4.06TSPIPLNMKK62 pKa = 10.3FEE64 pKa = 4.37GSINRR69 pKa = 11.84QLDD72 pKa = 3.36EE73 pKa = 5.5DD74 pKa = 4.6DD75 pKa = 5.54FIVSNGNIGNFEE87 pKa = 4.21RR88 pKa = 11.84GVPEE92 pKa = 3.91FTKK95 pKa = 10.52SRR97 pKa = 11.84EE98 pKa = 4.18DD99 pKa = 3.32IEE101 pKa = 4.47IEE103 pKa = 3.89NFQVEE108 pKa = 4.75GSAFEE113 pKa = 4.15IFIKK117 pKa = 10.66GISEE121 pKa = 3.99FLAEE125 pKa = 4.26EE126 pKa = 4.26FGDD129 pKa = 3.84AVISIAHH136 pKa = 4.66YY137 pKa = 9.68TKK139 pKa = 11.13DD140 pKa = 3.14NLFYY144 pKa = 10.84LSYY147 pKa = 10.97SNGVKK152 pKa = 10.25SLNLVTEE159 pKa = 4.82NKK161 pKa = 9.84QSTSITGDD169 pKa = 3.69DD170 pKa = 4.34LNSPLDD176 pKa = 3.6IEE178 pKa = 5.04ASLSGVDD185 pKa = 3.44QRR187 pKa = 11.84LYY189 pKa = 10.99LADD192 pKa = 3.77KK193 pKa = 10.73NKK195 pKa = 10.0QQVNVYY201 pKa = 10.67NNATVKK207 pKa = 10.73LDD209 pKa = 3.59DD210 pKa = 4.51FKK212 pKa = 11.82VPNYY216 pKa = 10.37DD217 pKa = 2.89SSNDD221 pKa = 3.48VGPTGIVTDD230 pKa = 4.5DD231 pKa = 4.3DD232 pKa = 4.52GNIYY236 pKa = 8.7VTVAFTPDD244 pKa = 2.63GTYY247 pKa = 10.64DD248 pKa = 3.4VEE250 pKa = 4.15YY251 pKa = 11.06DD252 pKa = 3.21KK253 pKa = 11.7VAVYY257 pKa = 10.47KK258 pKa = 10.37PGNYY262 pKa = 9.67NNPTVIPADD271 pKa = 3.56NSGLQLDD278 pKa = 4.21RR279 pKa = 11.84PYY281 pKa = 10.87RR282 pKa = 11.84IAVDD286 pKa = 3.45KK287 pKa = 11.09NGRR290 pKa = 11.84IYY292 pKa = 10.47IADD295 pKa = 4.0GGGSTPNGRR304 pKa = 11.84VLVFDD309 pKa = 4.0EE310 pKa = 5.03NYY312 pKa = 10.58NHH314 pKa = 6.73IHH316 pKa = 6.29TIEE319 pKa = 4.21GTQDD323 pKa = 3.06KK324 pKa = 10.26IGAPGSIIVDD334 pKa = 3.51DD335 pKa = 4.6FGYY338 pKa = 10.26IYY340 pKa = 10.16IIDD343 pKa = 4.06YY344 pKa = 11.19QNDD347 pKa = 2.91ITFNGIFQNPEE358 pKa = 4.03DD359 pKa = 4.78LLGEE363 pKa = 3.97YY364 pKa = 10.52DD365 pKa = 4.37KK366 pKa = 11.06IISTSYY372 pKa = 10.41SVNVYY377 pKa = 10.56DD378 pKa = 4.4SNANFEE384 pKa = 4.53YY385 pKa = 9.95VTEE388 pKa = 4.76FNQGLNLPIDD398 pKa = 4.15LEE400 pKa = 3.94IDD402 pKa = 3.44YY403 pKa = 10.98CGNILVNNLKK413 pKa = 10.7LSGPAPEE420 pKa = 4.5VLTGIDD426 pKa = 3.29INFDD430 pKa = 3.55FQLKK434 pKa = 8.48TFTRR438 pKa = 11.84HH439 pKa = 6.73DD440 pKa = 3.73NFTAEE445 pKa = 4.06VTPNGEE451 pKa = 3.99GLVEE455 pKa = 4.13VSLNPEE461 pKa = 3.96NEE463 pKa = 4.21FFPCDD468 pKa = 3.82PQPEE472 pKa = 4.37CSFSIEE478 pKa = 4.44YY479 pKa = 8.48KK480 pKa = 10.27TEE482 pKa = 4.02AAPDD486 pKa = 3.79PTFDD490 pKa = 4.58CPIEE494 pKa = 4.5SEE496 pKa = 4.25IDD498 pKa = 3.23PLFYY502 pKa = 11.16GNNCSVSVPDD512 pKa = 4.12YY513 pKa = 10.66EE514 pKa = 4.79ISNPQHH520 pKa = 6.38FEE522 pKa = 3.72SDD524 pKa = 3.24PFLVQTYY531 pKa = 8.11TEE533 pKa = 4.39NEE535 pKa = 3.9NSVSVLLKK543 pKa = 10.88VYY545 pKa = 10.58DD546 pKa = 4.03GEE548 pKa = 4.9NGDD551 pKa = 6.22LVDD554 pKa = 3.65EE555 pKa = 5.11CNFEE559 pKa = 4.78VDD561 pKa = 5.54LIDD564 pKa = 3.95NKK566 pKa = 11.14APVISDD572 pKa = 4.12CPPEE576 pKa = 4.45NIQYY580 pKa = 8.54TIQEE584 pKa = 4.67GEE586 pKa = 4.07THH588 pKa = 6.83ILDD591 pKa = 5.28DD592 pKa = 4.0YY593 pKa = 10.93TDD595 pKa = 3.77QISVNDD601 pKa = 3.85NCDD604 pKa = 3.62SNPTIVQDD612 pKa = 4.01PPAGTEE618 pKa = 3.53ITADD622 pKa = 3.36QSVTLIVTDD631 pKa = 3.62EE632 pKa = 4.4ADD634 pKa = 4.94NEE636 pKa = 4.61TEE638 pKa = 4.34CNFKK642 pKa = 11.16VEE644 pKa = 4.17ITEE647 pKa = 4.16VAVPDD652 pKa = 3.93PTFDD656 pKa = 4.36CPVEE660 pKa = 4.26NNLDD664 pKa = 3.69TIYY667 pKa = 11.3YY668 pKa = 10.37DD669 pKa = 4.71DD670 pKa = 3.97NCSYY674 pKa = 11.17NVPKK678 pKa = 10.88YY679 pKa = 10.55KK680 pKa = 10.17ISNPQHH686 pKa = 6.69FDD688 pKa = 3.33PEE690 pKa = 4.5PYY692 pKa = 10.4LIQSHH697 pKa = 5.72TEE699 pKa = 3.78NGDD702 pKa = 3.87SVSVTIKK709 pKa = 10.44VYY711 pKa = 10.42NGEE714 pKa = 3.96GGEE717 pKa = 4.46LVDD720 pKa = 3.66ICDD723 pKa = 3.99FEE725 pKa = 4.82VALEE729 pKa = 4.24DD730 pKa = 3.46NQAPVISGCPPEE742 pKa = 5.48NIQFTIQEE750 pKa = 4.64GEE752 pKa = 4.07THH754 pKa = 6.89ILDD757 pKa = 5.17DD758 pKa = 3.82YY759 pKa = 10.65TGQISVNDD767 pKa = 3.78NCDD770 pKa = 3.69SNPSIVQDD778 pKa = 3.89PPAGTEE784 pKa = 3.52ITADD788 pKa = 3.5RR789 pKa = 11.84LVTFTVTDD797 pKa = 3.48KK798 pKa = 11.44AGNEE802 pKa = 4.27TEE804 pKa = 4.35CNFMVEE810 pKa = 4.15ITEE813 pKa = 4.36EE814 pKa = 4.31EE815 pKa = 4.83PNTAPVAADD824 pKa = 3.19NTYY827 pKa = 10.84SIDD830 pKa = 3.61QNEE833 pKa = 4.76VLTIPAPGILQNDD846 pKa = 3.89TDD848 pKa = 4.79SEE850 pKa = 4.47DD851 pKa = 4.39DD852 pKa = 3.88NLTASLVTNVSNGTLTFNADD872 pKa = 2.77GSFTYY877 pKa = 10.09TPNTGFIGSDD887 pKa = 2.81SFTYY891 pKa = 10.03KK892 pKa = 10.96ANDD895 pKa = 3.75GEE897 pKa = 4.48LDD899 pKa = 3.87SNIATVTINVNSITAPNTAPVAADD923 pKa = 3.19NTYY926 pKa = 10.84SIDD929 pKa = 3.61QNEE932 pKa = 4.76VLTIPAPGILQNDD945 pKa = 3.79TDD947 pKa = 4.37SEE949 pKa = 4.33EE950 pKa = 4.91DD951 pKa = 3.53NLTASLDD958 pKa = 3.74SDD960 pKa = 4.02VSNGTLTFNADD971 pKa = 2.81GSFTYY976 pKa = 10.25IPNSDD981 pKa = 3.88FKK983 pKa = 11.34GQDD986 pKa = 2.92SFTYY990 pKa = 10.01RR991 pKa = 11.84ANDD994 pKa = 3.19GKK996 pKa = 10.8LYY998 pKa = 11.05SNIATVTITVNSTTAPNTAPVAVDD1022 pKa = 3.08NTYY1025 pKa = 10.91SIDD1028 pKa = 3.63QNEE1031 pKa = 4.76VLTIPAPGILQNDD1044 pKa = 3.79TDD1046 pKa = 4.37SEE1048 pKa = 4.33EE1049 pKa = 4.91DD1050 pKa = 3.53NLTASLDD1057 pKa = 3.74SDD1059 pKa = 4.02VSNGTLTLNADD1070 pKa = 3.4GSFTYY1075 pKa = 10.51EE1076 pKa = 3.87PDD1078 pKa = 3.54LGYY1081 pKa = 9.36TGQDD1085 pKa = 2.86SFTYY1089 pKa = 8.71TANDD1093 pKa = 3.69GTDD1096 pKa = 3.54DD1097 pKa = 4.37SNVATVTITVNSTTAPNTAPVAEE1120 pKa = 4.31NDD1122 pKa = 3.82TYY1124 pKa = 11.67AVDD1127 pKa = 3.58QDD1129 pKa = 3.75EE1130 pKa = 4.79VLPISSPGILQNDD1143 pKa = 3.43TDD1145 pKa = 4.47ADD1147 pKa = 4.12GDD1149 pKa = 4.01NLTASLVSDD1158 pKa = 3.92VSNGTLILNADD1169 pKa = 3.64GSFTYY1174 pKa = 9.84TPNSGYY1180 pKa = 10.62VGQDD1184 pKa = 2.61SFTYY1188 pKa = 8.73TANDD1192 pKa = 3.69GTDD1195 pKa = 3.54DD1196 pKa = 4.37SNVATVTITVNSTTAPNTAPVAEE1219 pKa = 4.31NDD1221 pKa = 3.82TYY1223 pKa = 11.67AVDD1226 pKa = 3.58QDD1228 pKa = 3.75EE1229 pKa = 4.79VLPISSPGILQNDD1242 pKa = 3.8TDD1244 pKa = 4.4SEE1246 pKa = 4.5GDD1248 pKa = 3.46NLTASLVSDD1257 pKa = 4.02VFNGTLTFNADD1268 pKa = 2.56GSFIYY1273 pKa = 10.34EE1274 pKa = 4.09PSSGFTGQDD1283 pKa = 2.78SFTYY1287 pKa = 10.04RR1288 pKa = 11.84ANDD1291 pKa = 3.35GKK1293 pKa = 10.88LDD1295 pKa = 4.06SNIATVTITVTKK1307 pKa = 9.91NDD1309 pKa = 3.7PAAPEE1314 pKa = 4.27INCLNHH1320 pKa = 7.08VIFLDD1325 pKa = 3.86KK1326 pKa = 10.87NGKK1329 pKa = 9.95ASLEE1333 pKa = 4.01PKK1335 pKa = 9.58TIYY1338 pKa = 10.85NGDD1341 pKa = 3.69RR1342 pKa = 11.84ADD1344 pKa = 4.78LEE1346 pKa = 4.82LKK1348 pKa = 10.7LSQTNFDD1355 pKa = 4.25CSNLGSNTVTLIATDD1370 pKa = 4.16PEE1372 pKa = 4.55TGLSSLCTAHH1382 pKa = 7.97IEE1384 pKa = 4.15VLDD1387 pKa = 4.36VISPEE1392 pKa = 3.85ASCISGYY1399 pKa = 8.59VLEE1402 pKa = 5.07LGEE1405 pKa = 5.77DD1406 pKa = 3.96GTASLAAAAIDD1417 pKa = 4.02NNSTDD1422 pKa = 2.93NCSIVSRR1429 pKa = 11.84SVSRR1433 pKa = 11.84QNFTRR1438 pKa = 11.84DD1439 pKa = 3.54DD1440 pKa = 3.64IGDD1443 pKa = 3.44VSVRR1447 pKa = 11.84LTVRR1451 pKa = 11.84DD1452 pKa = 3.29AAGNEE1457 pKa = 4.55GSCEE1461 pKa = 4.04SVIKK1465 pKa = 11.04VIDD1468 pKa = 3.64KK1469 pKa = 8.05QTSDD1473 pKa = 3.81FQCKK1477 pKa = 9.17EE1478 pKa = 4.01KK1479 pKa = 11.19VSVSLNEE1486 pKa = 4.1NGNATLAVEE1495 pKa = 4.17EE1496 pKa = 5.69LYY1498 pKa = 10.78TGDD1501 pKa = 3.37ASGINFDD1508 pKa = 3.83KK1509 pKa = 11.22NNYY1512 pKa = 8.64QFSCADD1518 pKa = 4.12LGTKK1522 pKa = 8.9TIQFNYY1528 pKa = 9.41SGRR1531 pKa = 11.84IEE1533 pKa = 4.36GSCSIKK1539 pKa = 10.8VEE1541 pKa = 4.43VKK1543 pKa = 10.89DD1544 pKa = 4.18EE1545 pKa = 4.07LAPTINTNLINASLDD1560 pKa = 3.44EE1561 pKa = 5.42DD1562 pKa = 4.05GFFYY1566 pKa = 10.93LDD1568 pKa = 3.53EE1569 pKa = 4.51NRR1571 pKa = 11.84VEE1573 pKa = 5.95AEE1575 pKa = 4.36DD1576 pKa = 3.65NCNTPLQITLEE1587 pKa = 4.24KK1588 pKa = 10.3TVFTCEE1594 pKa = 4.65DD1595 pKa = 3.2VGQNSILVEE1604 pKa = 4.2AEE1606 pKa = 3.83DD1607 pKa = 3.76ASGIKK1612 pKa = 8.92TSKK1615 pKa = 9.92EE1616 pKa = 3.21ITVIVSGAPCEE1627 pKa = 4.01PSIEE1631 pKa = 4.23FNDD1634 pKa = 3.48IRR1636 pKa = 11.84ISPNPSAGIFKK1647 pKa = 10.37IGTPTDD1653 pKa = 3.1IKK1655 pKa = 10.66INEE1658 pKa = 4.17VKK1660 pKa = 10.88VFDD1663 pKa = 3.51SRR1665 pKa = 11.84GRR1667 pKa = 11.84FITQKK1672 pKa = 10.35DD1673 pKa = 3.74YY1674 pKa = 11.84NMNFRR1679 pKa = 11.84FYY1681 pKa = 11.22KK1682 pKa = 10.74LNLEE1686 pKa = 4.24SLQEE1690 pKa = 4.02SVYY1693 pKa = 10.22TLQIFTNNGEE1703 pKa = 4.07FVKK1706 pKa = 10.7RR1707 pKa = 11.84IIIKK1711 pKa = 10.21RR1712 pKa = 3.59
MM1 pKa = 7.35NKK3 pKa = 9.44YY4 pKa = 10.05HH5 pKa = 6.93SLPTGPNFRR14 pKa = 11.84YY15 pKa = 9.57LPKK18 pKa = 10.57GLYY21 pKa = 8.97PLLFIFFFLLKK32 pKa = 10.29SGVGFGQEE40 pKa = 3.65NCIITSTATDD50 pKa = 3.7PTEE53 pKa = 4.06TSPIPLNMKK62 pKa = 10.3FEE64 pKa = 4.37GSINRR69 pKa = 11.84QLDD72 pKa = 3.36EE73 pKa = 5.5DD74 pKa = 4.6DD75 pKa = 5.54FIVSNGNIGNFEE87 pKa = 4.21RR88 pKa = 11.84GVPEE92 pKa = 3.91FTKK95 pKa = 10.52SRR97 pKa = 11.84EE98 pKa = 4.18DD99 pKa = 3.32IEE101 pKa = 4.47IEE103 pKa = 3.89NFQVEE108 pKa = 4.75GSAFEE113 pKa = 4.15IFIKK117 pKa = 10.66GISEE121 pKa = 3.99FLAEE125 pKa = 4.26EE126 pKa = 4.26FGDD129 pKa = 3.84AVISIAHH136 pKa = 4.66YY137 pKa = 9.68TKK139 pKa = 11.13DD140 pKa = 3.14NLFYY144 pKa = 10.84LSYY147 pKa = 10.97SNGVKK152 pKa = 10.25SLNLVTEE159 pKa = 4.82NKK161 pKa = 9.84QSTSITGDD169 pKa = 3.69DD170 pKa = 4.34LNSPLDD176 pKa = 3.6IEE178 pKa = 5.04ASLSGVDD185 pKa = 3.44QRR187 pKa = 11.84LYY189 pKa = 10.99LADD192 pKa = 3.77KK193 pKa = 10.73NKK195 pKa = 10.0QQVNVYY201 pKa = 10.67NNATVKK207 pKa = 10.73LDD209 pKa = 3.59DD210 pKa = 4.51FKK212 pKa = 11.82VPNYY216 pKa = 10.37DD217 pKa = 2.89SSNDD221 pKa = 3.48VGPTGIVTDD230 pKa = 4.5DD231 pKa = 4.3DD232 pKa = 4.52GNIYY236 pKa = 8.7VTVAFTPDD244 pKa = 2.63GTYY247 pKa = 10.64DD248 pKa = 3.4VEE250 pKa = 4.15YY251 pKa = 11.06DD252 pKa = 3.21KK253 pKa = 11.7VAVYY257 pKa = 10.47KK258 pKa = 10.37PGNYY262 pKa = 9.67NNPTVIPADD271 pKa = 3.56NSGLQLDD278 pKa = 4.21RR279 pKa = 11.84PYY281 pKa = 10.87RR282 pKa = 11.84IAVDD286 pKa = 3.45KK287 pKa = 11.09NGRR290 pKa = 11.84IYY292 pKa = 10.47IADD295 pKa = 4.0GGGSTPNGRR304 pKa = 11.84VLVFDD309 pKa = 4.0EE310 pKa = 5.03NYY312 pKa = 10.58NHH314 pKa = 6.73IHH316 pKa = 6.29TIEE319 pKa = 4.21GTQDD323 pKa = 3.06KK324 pKa = 10.26IGAPGSIIVDD334 pKa = 3.51DD335 pKa = 4.6FGYY338 pKa = 10.26IYY340 pKa = 10.16IIDD343 pKa = 4.06YY344 pKa = 11.19QNDD347 pKa = 2.91ITFNGIFQNPEE358 pKa = 4.03DD359 pKa = 4.78LLGEE363 pKa = 3.97YY364 pKa = 10.52DD365 pKa = 4.37KK366 pKa = 11.06IISTSYY372 pKa = 10.41SVNVYY377 pKa = 10.56DD378 pKa = 4.4SNANFEE384 pKa = 4.53YY385 pKa = 9.95VTEE388 pKa = 4.76FNQGLNLPIDD398 pKa = 4.15LEE400 pKa = 3.94IDD402 pKa = 3.44YY403 pKa = 10.98CGNILVNNLKK413 pKa = 10.7LSGPAPEE420 pKa = 4.5VLTGIDD426 pKa = 3.29INFDD430 pKa = 3.55FQLKK434 pKa = 8.48TFTRR438 pKa = 11.84HH439 pKa = 6.73DD440 pKa = 3.73NFTAEE445 pKa = 4.06VTPNGEE451 pKa = 3.99GLVEE455 pKa = 4.13VSLNPEE461 pKa = 3.96NEE463 pKa = 4.21FFPCDD468 pKa = 3.82PQPEE472 pKa = 4.37CSFSIEE478 pKa = 4.44YY479 pKa = 8.48KK480 pKa = 10.27TEE482 pKa = 4.02AAPDD486 pKa = 3.79PTFDD490 pKa = 4.58CPIEE494 pKa = 4.5SEE496 pKa = 4.25IDD498 pKa = 3.23PLFYY502 pKa = 11.16GNNCSVSVPDD512 pKa = 4.12YY513 pKa = 10.66EE514 pKa = 4.79ISNPQHH520 pKa = 6.38FEE522 pKa = 3.72SDD524 pKa = 3.24PFLVQTYY531 pKa = 8.11TEE533 pKa = 4.39NEE535 pKa = 3.9NSVSVLLKK543 pKa = 10.88VYY545 pKa = 10.58DD546 pKa = 4.03GEE548 pKa = 4.9NGDD551 pKa = 6.22LVDD554 pKa = 3.65EE555 pKa = 5.11CNFEE559 pKa = 4.78VDD561 pKa = 5.54LIDD564 pKa = 3.95NKK566 pKa = 11.14APVISDD572 pKa = 4.12CPPEE576 pKa = 4.45NIQYY580 pKa = 8.54TIQEE584 pKa = 4.67GEE586 pKa = 4.07THH588 pKa = 6.83ILDD591 pKa = 5.28DD592 pKa = 4.0YY593 pKa = 10.93TDD595 pKa = 3.77QISVNDD601 pKa = 3.85NCDD604 pKa = 3.62SNPTIVQDD612 pKa = 4.01PPAGTEE618 pKa = 3.53ITADD622 pKa = 3.36QSVTLIVTDD631 pKa = 3.62EE632 pKa = 4.4ADD634 pKa = 4.94NEE636 pKa = 4.61TEE638 pKa = 4.34CNFKK642 pKa = 11.16VEE644 pKa = 4.17ITEE647 pKa = 4.16VAVPDD652 pKa = 3.93PTFDD656 pKa = 4.36CPVEE660 pKa = 4.26NNLDD664 pKa = 3.69TIYY667 pKa = 11.3YY668 pKa = 10.37DD669 pKa = 4.71DD670 pKa = 3.97NCSYY674 pKa = 11.17NVPKK678 pKa = 10.88YY679 pKa = 10.55KK680 pKa = 10.17ISNPQHH686 pKa = 6.69FDD688 pKa = 3.33PEE690 pKa = 4.5PYY692 pKa = 10.4LIQSHH697 pKa = 5.72TEE699 pKa = 3.78NGDD702 pKa = 3.87SVSVTIKK709 pKa = 10.44VYY711 pKa = 10.42NGEE714 pKa = 3.96GGEE717 pKa = 4.46LVDD720 pKa = 3.66ICDD723 pKa = 3.99FEE725 pKa = 4.82VALEE729 pKa = 4.24DD730 pKa = 3.46NQAPVISGCPPEE742 pKa = 5.48NIQFTIQEE750 pKa = 4.64GEE752 pKa = 4.07THH754 pKa = 6.89ILDD757 pKa = 5.17DD758 pKa = 3.82YY759 pKa = 10.65TGQISVNDD767 pKa = 3.78NCDD770 pKa = 3.69SNPSIVQDD778 pKa = 3.89PPAGTEE784 pKa = 3.52ITADD788 pKa = 3.5RR789 pKa = 11.84LVTFTVTDD797 pKa = 3.48KK798 pKa = 11.44AGNEE802 pKa = 4.27TEE804 pKa = 4.35CNFMVEE810 pKa = 4.15ITEE813 pKa = 4.36EE814 pKa = 4.31EE815 pKa = 4.83PNTAPVAADD824 pKa = 3.19NTYY827 pKa = 10.84SIDD830 pKa = 3.61QNEE833 pKa = 4.76VLTIPAPGILQNDD846 pKa = 3.89TDD848 pKa = 4.79SEE850 pKa = 4.47DD851 pKa = 4.39DD852 pKa = 3.88NLTASLVTNVSNGTLTFNADD872 pKa = 2.77GSFTYY877 pKa = 10.09TPNTGFIGSDD887 pKa = 2.81SFTYY891 pKa = 10.03KK892 pKa = 10.96ANDD895 pKa = 3.75GEE897 pKa = 4.48LDD899 pKa = 3.87SNIATVTINVNSITAPNTAPVAADD923 pKa = 3.19NTYY926 pKa = 10.84SIDD929 pKa = 3.61QNEE932 pKa = 4.76VLTIPAPGILQNDD945 pKa = 3.79TDD947 pKa = 4.37SEE949 pKa = 4.33EE950 pKa = 4.91DD951 pKa = 3.53NLTASLDD958 pKa = 3.74SDD960 pKa = 4.02VSNGTLTFNADD971 pKa = 2.81GSFTYY976 pKa = 10.25IPNSDD981 pKa = 3.88FKK983 pKa = 11.34GQDD986 pKa = 2.92SFTYY990 pKa = 10.01RR991 pKa = 11.84ANDD994 pKa = 3.19GKK996 pKa = 10.8LYY998 pKa = 11.05SNIATVTITVNSTTAPNTAPVAVDD1022 pKa = 3.08NTYY1025 pKa = 10.91SIDD1028 pKa = 3.63QNEE1031 pKa = 4.76VLTIPAPGILQNDD1044 pKa = 3.79TDD1046 pKa = 4.37SEE1048 pKa = 4.33EE1049 pKa = 4.91DD1050 pKa = 3.53NLTASLDD1057 pKa = 3.74SDD1059 pKa = 4.02VSNGTLTLNADD1070 pKa = 3.4GSFTYY1075 pKa = 10.51EE1076 pKa = 3.87PDD1078 pKa = 3.54LGYY1081 pKa = 9.36TGQDD1085 pKa = 2.86SFTYY1089 pKa = 8.71TANDD1093 pKa = 3.69GTDD1096 pKa = 3.54DD1097 pKa = 4.37SNVATVTITVNSTTAPNTAPVAEE1120 pKa = 4.31NDD1122 pKa = 3.82TYY1124 pKa = 11.67AVDD1127 pKa = 3.58QDD1129 pKa = 3.75EE1130 pKa = 4.79VLPISSPGILQNDD1143 pKa = 3.43TDD1145 pKa = 4.47ADD1147 pKa = 4.12GDD1149 pKa = 4.01NLTASLVSDD1158 pKa = 3.92VSNGTLILNADD1169 pKa = 3.64GSFTYY1174 pKa = 9.84TPNSGYY1180 pKa = 10.62VGQDD1184 pKa = 2.61SFTYY1188 pKa = 8.73TANDD1192 pKa = 3.69GTDD1195 pKa = 3.54DD1196 pKa = 4.37SNVATVTITVNSTTAPNTAPVAEE1219 pKa = 4.31NDD1221 pKa = 3.82TYY1223 pKa = 11.67AVDD1226 pKa = 3.58QDD1228 pKa = 3.75EE1229 pKa = 4.79VLPISSPGILQNDD1242 pKa = 3.8TDD1244 pKa = 4.4SEE1246 pKa = 4.5GDD1248 pKa = 3.46NLTASLVSDD1257 pKa = 4.02VFNGTLTFNADD1268 pKa = 2.56GSFIYY1273 pKa = 10.34EE1274 pKa = 4.09PSSGFTGQDD1283 pKa = 2.78SFTYY1287 pKa = 10.04RR1288 pKa = 11.84ANDD1291 pKa = 3.35GKK1293 pKa = 10.88LDD1295 pKa = 4.06SNIATVTITVTKK1307 pKa = 9.91NDD1309 pKa = 3.7PAAPEE1314 pKa = 4.27INCLNHH1320 pKa = 7.08VIFLDD1325 pKa = 3.86KK1326 pKa = 10.87NGKK1329 pKa = 9.95ASLEE1333 pKa = 4.01PKK1335 pKa = 9.58TIYY1338 pKa = 10.85NGDD1341 pKa = 3.69RR1342 pKa = 11.84ADD1344 pKa = 4.78LEE1346 pKa = 4.82LKK1348 pKa = 10.7LSQTNFDD1355 pKa = 4.25CSNLGSNTVTLIATDD1370 pKa = 4.16PEE1372 pKa = 4.55TGLSSLCTAHH1382 pKa = 7.97IEE1384 pKa = 4.15VLDD1387 pKa = 4.36VISPEE1392 pKa = 3.85ASCISGYY1399 pKa = 8.59VLEE1402 pKa = 5.07LGEE1405 pKa = 5.77DD1406 pKa = 3.96GTASLAAAAIDD1417 pKa = 4.02NNSTDD1422 pKa = 2.93NCSIVSRR1429 pKa = 11.84SVSRR1433 pKa = 11.84QNFTRR1438 pKa = 11.84DD1439 pKa = 3.54DD1440 pKa = 3.64IGDD1443 pKa = 3.44VSVRR1447 pKa = 11.84LTVRR1451 pKa = 11.84DD1452 pKa = 3.29AAGNEE1457 pKa = 4.55GSCEE1461 pKa = 4.04SVIKK1465 pKa = 11.04VIDD1468 pKa = 3.64KK1469 pKa = 8.05QTSDD1473 pKa = 3.81FQCKK1477 pKa = 9.17EE1478 pKa = 4.01KK1479 pKa = 11.19VSVSLNEE1486 pKa = 4.1NGNATLAVEE1495 pKa = 4.17EE1496 pKa = 5.69LYY1498 pKa = 10.78TGDD1501 pKa = 3.37ASGINFDD1508 pKa = 3.83KK1509 pKa = 11.22NNYY1512 pKa = 8.64QFSCADD1518 pKa = 4.12LGTKK1522 pKa = 8.9TIQFNYY1528 pKa = 9.41SGRR1531 pKa = 11.84IEE1533 pKa = 4.36GSCSIKK1539 pKa = 10.8VEE1541 pKa = 4.43VKK1543 pKa = 10.89DD1544 pKa = 4.18EE1545 pKa = 4.07LAPTINTNLINASLDD1560 pKa = 3.44EE1561 pKa = 5.42DD1562 pKa = 4.05GFFYY1566 pKa = 10.93LDD1568 pKa = 3.53EE1569 pKa = 4.51NRR1571 pKa = 11.84VEE1573 pKa = 5.95AEE1575 pKa = 4.36DD1576 pKa = 3.65NCNTPLQITLEE1587 pKa = 4.24KK1588 pKa = 10.3TVFTCEE1594 pKa = 4.65DD1595 pKa = 3.2VGQNSILVEE1604 pKa = 4.2AEE1606 pKa = 3.83DD1607 pKa = 3.76ASGIKK1612 pKa = 8.92TSKK1615 pKa = 9.92EE1616 pKa = 3.21ITVIVSGAPCEE1627 pKa = 4.01PSIEE1631 pKa = 4.23FNDD1634 pKa = 3.48IRR1636 pKa = 11.84ISPNPSAGIFKK1647 pKa = 10.37IGTPTDD1653 pKa = 3.1IKK1655 pKa = 10.66INEE1658 pKa = 4.17VKK1660 pKa = 10.88VFDD1663 pKa = 3.51SRR1665 pKa = 11.84GRR1667 pKa = 11.84FITQKK1672 pKa = 10.35DD1673 pKa = 3.74YY1674 pKa = 11.84NMNFRR1679 pKa = 11.84FYY1681 pKa = 11.22KK1682 pKa = 10.74LNLEE1686 pKa = 4.24SLQEE1690 pKa = 4.02SVYY1693 pKa = 10.22TLQIFTNNGEE1703 pKa = 4.07FVKK1706 pKa = 10.7RR1707 pKa = 11.84IIIKK1711 pKa = 10.21RR1712 pKa = 3.59
Molecular weight: 186.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5C4P9|A0A1M5C4P9_9FLAO Probable transaldolase OS=Salegentibacter echinorum OX=1073325 GN=tal PE=3 SV=1
MM1 pKa = 7.12FQFDD5 pKa = 3.71QEE7 pKa = 4.27TWVYY11 pKa = 10.11IILIAVFLAYY21 pKa = 9.27FIWNSRR27 pKa = 11.84RR28 pKa = 11.84AKK30 pKa = 10.25KK31 pKa = 9.86NRR33 pKa = 11.84KK34 pKa = 8.15ARR36 pKa = 11.84KK37 pKa = 8.53NRR39 pKa = 11.84NFRR42 pKa = 11.84KK43 pKa = 9.61RR44 pKa = 11.84YY45 pKa = 6.27MEE47 pKa = 4.0RR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.11EE51 pKa = 3.91QDD53 pKa = 2.89KK54 pKa = 9.47TT55 pKa = 3.68
MM1 pKa = 7.12FQFDD5 pKa = 3.71QEE7 pKa = 4.27TWVYY11 pKa = 10.11IILIAVFLAYY21 pKa = 9.27FIWNSRR27 pKa = 11.84RR28 pKa = 11.84AKK30 pKa = 10.25KK31 pKa = 9.86NRR33 pKa = 11.84KK34 pKa = 8.15ARR36 pKa = 11.84KK37 pKa = 8.53NRR39 pKa = 11.84NFRR42 pKa = 11.84KK43 pKa = 9.61RR44 pKa = 11.84YY45 pKa = 6.27MEE47 pKa = 4.0RR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.11EE51 pKa = 3.91QDD53 pKa = 2.89KK54 pKa = 9.47TT55 pKa = 3.68
Molecular weight: 7.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1135832 |
29 |
2388 |
326.5 |
36.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.598 ± 0.034 | 0.665 ± 0.012 |
5.459 ± 0.034 | 7.459 ± 0.042 |
5.237 ± 0.03 | 6.415 ± 0.039 |
1.766 ± 0.019 | 7.579 ± 0.039 |
8.208 ± 0.05 | 9.466 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.22 ± 0.021 | 5.895 ± 0.039 |
3.464 ± 0.021 | 3.422 ± 0.025 |
3.812 ± 0.025 | 6.229 ± 0.034 |
5.151 ± 0.029 | 5.909 ± 0.03 |
1.073 ± 0.014 | 3.976 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
