
Lewinellaceae bacterium SD302
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; unclassified Lewinellaceae
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
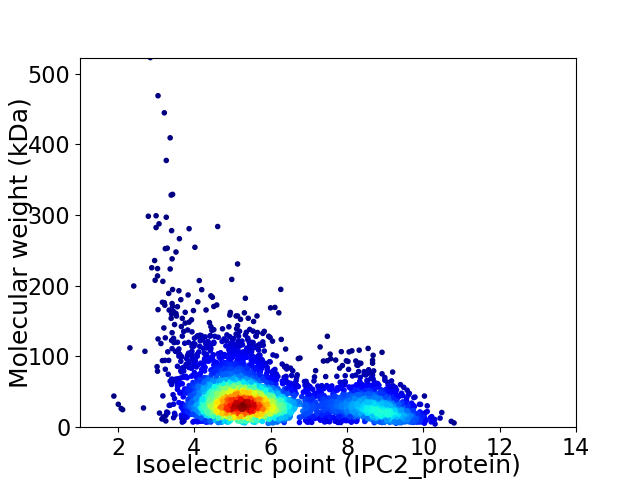
Virtual 2D-PAGE plot for 4052 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
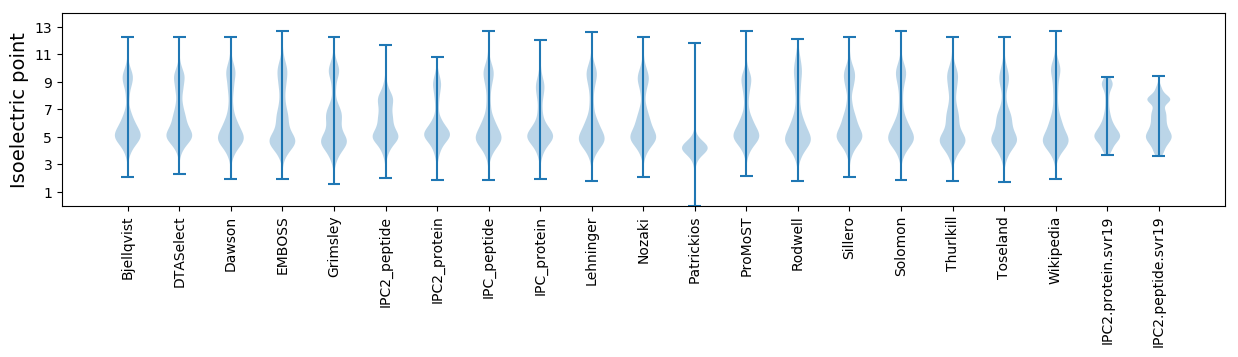
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C6CQR1|A0A2C6CQR1_9BACT Uncharacterized protein OS=Lewinellaceae bacterium SD302 OX=2014804 GN=CEQ90_07730 PE=4 SV=1
MM1 pKa = 7.38RR2 pKa = 11.84TLTTFLFFLAALNFIAGQSFRR23 pKa = 11.84TTLVSPATVSRR34 pKa = 11.84QTSNSDD40 pKa = 3.45LNGDD44 pKa = 3.94GHH46 pKa = 7.0QDD48 pKa = 4.08LITVNGEE55 pKa = 4.0VGWLKK60 pKa = 10.32WVVGAHH66 pKa = 7.41AYY68 pKa = 9.6PEE70 pKa = 4.14YY71 pKa = 10.89RR72 pKa = 11.84KK73 pKa = 8.39ITPDD77 pKa = 2.99EE78 pKa = 4.58NYY80 pKa = 8.93THH82 pKa = 7.45ALTGDD87 pKa = 3.33VDD89 pKa = 4.15SDD91 pKa = 3.77GDD93 pKa = 3.6IDD95 pKa = 4.92IVALGDD101 pKa = 3.73DD102 pKa = 3.63RR103 pKa = 11.84VVLLKK108 pKa = 11.28NNDD111 pKa = 3.28GEE113 pKa = 4.36AGFWLEE119 pKa = 5.07SMIDD123 pKa = 3.72DD124 pKa = 4.9EE125 pKa = 5.35EE126 pKa = 5.16VNDD129 pKa = 4.85PDD131 pKa = 3.5WGALGDD137 pKa = 3.74IDD139 pKa = 5.83GDD141 pKa = 3.91GDD143 pKa = 4.68LDD145 pKa = 4.24LLVASRR151 pKa = 11.84YY152 pKa = 9.87GYY154 pKa = 10.31YY155 pKa = 10.41LGWYY159 pKa = 7.79EE160 pKa = 4.11NTDD163 pKa = 3.24GTGEE167 pKa = 4.0FQFYY171 pKa = 11.32AEE173 pKa = 5.63LPDD176 pKa = 3.88AQNARR181 pKa = 11.84SFGALVDD188 pKa = 4.38FDD190 pKa = 4.04QDD192 pKa = 2.76GDD194 pKa = 4.45LDD196 pKa = 4.04VLSAGCYY203 pKa = 9.8LDD205 pKa = 5.5CTLEE209 pKa = 3.92WYY211 pKa = 9.0EE212 pKa = 3.99NLDD215 pKa = 3.55GEE217 pKa = 5.01GEE219 pKa = 3.95FGIRR223 pKa = 11.84KK224 pKa = 9.39EE225 pKa = 4.7IYY227 pKa = 9.23FDD229 pKa = 4.33DD230 pKa = 4.25YY231 pKa = 11.48GIEE234 pKa = 4.02NMVIDD239 pKa = 4.31VGDD242 pKa = 3.56VDD244 pKa = 3.89GDD246 pKa = 4.0GYY248 pKa = 11.02EE249 pKa = 5.63DD250 pKa = 4.12IMTTSTYY257 pKa = 10.17YY258 pKa = 10.7KK259 pKa = 9.6VIRR262 pKa = 11.84WHH264 pKa = 6.71RR265 pKa = 11.84NLIGSGEE272 pKa = 4.17MGFAAAVDD280 pKa = 3.36ISDD283 pKa = 4.19FDD285 pKa = 3.62WARR288 pKa = 11.84SIEE291 pKa = 4.38LTDD294 pKa = 4.74LDD296 pKa = 4.61NDD298 pKa = 3.48NDD300 pKa = 3.96LDD302 pKa = 3.81IAVAGDD308 pKa = 4.5DD309 pKa = 3.89DD310 pKa = 3.9THH312 pKa = 8.55LIGWYY317 pKa = 9.21EE318 pKa = 3.92NWSGDD323 pKa = 4.0GLFSNLNIIDD333 pKa = 4.78DD334 pKa = 4.31EE335 pKa = 4.49TDD337 pKa = 2.62VDD339 pKa = 4.29YY340 pKa = 11.71SRR342 pKa = 11.84LICFDD347 pKa = 2.99IDD349 pKa = 3.69EE350 pKa = 5.36NGTQDD355 pKa = 3.31ILTAGTLNSHH365 pKa = 6.57TIGWYY370 pKa = 10.37ANEE373 pKa = 4.25EE374 pKa = 4.18NTGSFGRR381 pKa = 11.84VNEE384 pKa = 4.1LVHH387 pKa = 6.75PYY389 pKa = 10.04IADD392 pKa = 4.05EE393 pKa = 4.25IMLNDD398 pKa = 3.28VDD400 pKa = 5.41GDD402 pKa = 4.17GNTDD406 pKa = 2.95IVAGGSWYY414 pKa = 9.75TNSGYY419 pKa = 10.93DD420 pKa = 3.33YY421 pKa = 11.09LAFSNIRR428 pKa = 11.84MVDD431 pKa = 3.66EE432 pKa = 4.91EE433 pKa = 4.55GHH435 pKa = 6.29LSLVEE440 pKa = 4.9DD441 pKa = 4.44FTNDD445 pKa = 3.94GITDD449 pKa = 5.94LITLEE454 pKa = 4.16RR455 pKa = 11.84LPLSTHH461 pKa = 5.78KK462 pKa = 10.79KK463 pKa = 7.45IVLYY467 pKa = 10.17EE468 pKa = 3.84RR469 pKa = 11.84SGEE472 pKa = 4.0QFIFSSDD479 pKa = 2.77IVEE482 pKa = 4.15VDD484 pKa = 2.97NASIKK489 pKa = 10.88NLVSLDD495 pKa = 3.56YY496 pKa = 11.56DD497 pKa = 3.56NDD499 pKa = 3.75GDD501 pKa = 5.17LDD503 pKa = 3.96FVYY506 pKa = 9.05QTAYY510 pKa = 10.8DD511 pKa = 4.14VVWLCEE517 pKa = 4.0NLGSDD522 pKa = 3.77NGFAAPVEE530 pKa = 4.24IADD533 pKa = 4.24TEE535 pKa = 4.38VSVNNFEE542 pKa = 5.91LGDD545 pKa = 4.1LDD547 pKa = 3.78GDD549 pKa = 4.31GYY551 pKa = 11.74VDD553 pKa = 4.43ILIGTDD559 pKa = 2.53WNQRR563 pKa = 11.84MGWYY567 pKa = 9.96RR568 pKa = 11.84NEE570 pKa = 4.27AGSGTFSDD578 pKa = 4.39LAIISSGQPYY588 pKa = 9.68EE589 pKa = 3.95QLDD592 pKa = 3.51WDD594 pKa = 4.49DD595 pKa = 3.77QDD597 pKa = 5.26GDD599 pKa = 3.78GDD601 pKa = 4.46LDD603 pKa = 3.75LVYY606 pKa = 10.71VYY608 pKa = 10.6EE609 pKa = 4.19EE610 pKa = 4.28NILRR614 pKa = 11.84RR615 pKa = 11.84NNEE618 pKa = 3.7GNGVFSSEE626 pKa = 3.79EE627 pKa = 4.01VVYY630 pKa = 10.63HH631 pKa = 6.84DD632 pKa = 4.19PAGGHH637 pKa = 4.9SFRR640 pKa = 11.84RR641 pKa = 11.84IYY643 pKa = 11.16CNDD646 pKa = 3.35HH647 pKa = 6.74NDD649 pKa = 3.43DD650 pKa = 3.82GLRR653 pKa = 11.84DD654 pKa = 3.83FLFFDD659 pKa = 3.36QSSYY663 pKa = 9.4GWRR666 pKa = 11.84FLTAEE671 pKa = 3.97IEE673 pKa = 4.08GGYY676 pKa = 10.12LLSAEE681 pKa = 4.6AEE683 pKa = 4.52FIPSNNVEE691 pKa = 4.37PIKK694 pKa = 10.72FYY696 pKa = 11.3DD697 pKa = 4.24LDD699 pKa = 4.09NDD701 pKa = 4.32GDD703 pKa = 4.43DD704 pKa = 5.75DD705 pKa = 4.09MLFAQGDD712 pKa = 3.8KK713 pKa = 11.03LYY715 pKa = 11.17VSEE718 pKa = 4.65NLNSGKK724 pKa = 9.89VLQGLIYY731 pKa = 10.51YY732 pKa = 9.99DD733 pKa = 3.86EE734 pKa = 4.77NEE736 pKa = 4.11NGINDD741 pKa = 4.35DD742 pKa = 3.78SDD744 pKa = 4.24SPLVDD749 pKa = 3.17QLVSIDD755 pKa = 3.53PTQLFLHH762 pKa = 6.72SNTDD766 pKa = 3.5GLFSTVLGNGQYY778 pKa = 9.22EE779 pKa = 4.58VRR781 pKa = 11.84CDD783 pKa = 4.38PEE785 pKa = 5.13DD786 pKa = 3.16GWGFVGDD793 pKa = 4.54SIVNIQTSSDD803 pKa = 3.28ADD805 pKa = 4.31TIWVEE810 pKa = 4.24FPLINTDD817 pKa = 3.17TTSRR821 pKa = 11.84PSVEE825 pKa = 4.26VTSGPVRR832 pKa = 11.84CGFEE836 pKa = 4.09VPFWLTVNNDD846 pKa = 2.99GYY848 pKa = 11.59VPEE851 pKa = 4.57TGDD854 pKa = 3.34FYY856 pKa = 11.33FYY858 pKa = 10.97LDD860 pKa = 3.99SLVALAIDD868 pKa = 4.42PADD871 pKa = 3.95HH872 pKa = 7.18PNLFVDD878 pKa = 3.82AEE880 pKa = 4.31GRR882 pKa = 11.84TGLFWEE888 pKa = 5.12QIDD891 pKa = 3.78ISPGARR897 pKa = 11.84WQTQVLLQMPSVDD910 pKa = 5.05FIGEE914 pKa = 3.84ALGFNGIFEE923 pKa = 4.49YY924 pKa = 10.86AVGINEE930 pKa = 4.03LQSNGEE936 pKa = 3.88YY937 pKa = 10.27TRR939 pKa = 11.84VLNCSYY945 pKa = 11.14DD946 pKa = 3.75PNDD949 pKa = 3.54KK950 pKa = 10.75SVEE953 pKa = 4.16EE954 pKa = 4.42NLPGYY959 pKa = 9.16PGYY962 pKa = 11.73SMVGDD967 pKa = 3.65SLEE970 pKa = 3.89YY971 pKa = 10.62LIRR974 pKa = 11.84FQNLGSDD981 pKa = 3.33TAINIKK987 pKa = 10.46VSDD990 pKa = 3.8QLSEE994 pKa = 3.95HH995 pKa = 6.74FDD997 pKa = 3.39RR998 pKa = 11.84SKK1000 pKa = 11.32LRR1002 pKa = 11.84ILSASHH1008 pKa = 6.94NYY1010 pKa = 10.07RR1011 pKa = 11.84MVLTDD1016 pKa = 4.08DD1017 pKa = 3.66NRR1019 pKa = 11.84LNVYY1023 pKa = 9.93FDD1025 pKa = 4.5NIYY1028 pKa = 10.86LPDD1031 pKa = 3.99AMTDD1035 pKa = 3.29EE1036 pKa = 5.05AGSHH1040 pKa = 5.74GFIKK1044 pKa = 10.39FRR1046 pKa = 11.84VGMLPGSTDD1055 pKa = 2.85GDD1057 pKa = 3.96VIEE1060 pKa = 4.8NQADD1064 pKa = 3.42IYY1066 pKa = 11.19FDD1068 pKa = 4.28FNPPITTNVVYY1079 pKa = 8.95NTILEE1084 pKa = 4.14RR1085 pKa = 11.84LIYY1088 pKa = 8.85PAQILAPKK1096 pKa = 9.98CFEE1099 pKa = 4.35FSDD1102 pKa = 4.25GSISVEE1108 pKa = 3.81EE1109 pKa = 4.21VVNGVDD1115 pKa = 3.84YY1116 pKa = 10.13FWNGISSSNHH1126 pKa = 5.54IEE1128 pKa = 4.02NLSAGTNTLEE1138 pKa = 4.7VYY1140 pKa = 10.56RR1141 pKa = 11.84SDD1143 pKa = 4.07GKK1145 pKa = 10.7ILADD1149 pKa = 3.4TSYY1152 pKa = 11.14QITDD1156 pKa = 3.58PDD1158 pKa = 3.64ILEE1161 pKa = 4.06VTAEE1165 pKa = 3.97IEE1167 pKa = 4.05PASFANSGSIIATPTGGTAPYY1188 pKa = 9.4TYY1190 pKa = 10.29VWQQFPEE1197 pKa = 4.07QQGNEE1202 pKa = 4.09IGDD1205 pKa = 4.08LAAGNYY1211 pKa = 9.09DD1212 pKa = 4.01LQVIDD1217 pKa = 5.18ANGCIIDD1224 pKa = 3.86AVFIVPEE1231 pKa = 4.21VVSTAEE1237 pKa = 4.25HH1238 pKa = 6.47LAKK1241 pKa = 10.66SPLSLRR1247 pKa = 11.84IYY1249 pKa = 9.3PNPARR1254 pKa = 11.84ALEE1257 pKa = 4.05FVNIRR1262 pKa = 11.84FSEE1265 pKa = 4.29VKK1267 pKa = 10.55NEE1269 pKa = 4.07SEE1271 pKa = 4.38LLVYY1275 pKa = 10.83DD1276 pKa = 3.47NLKK1279 pKa = 10.2RR1280 pKa = 11.84IVVRR1284 pKa = 11.84RR1285 pKa = 11.84SIAPGSYY1292 pKa = 10.29EE1293 pKa = 4.13LTDD1296 pKa = 4.11LKK1298 pKa = 10.69LAPGNYY1304 pKa = 9.27FLVLISGGKK1313 pKa = 8.11HH1314 pKa = 4.32ALGRR1318 pKa = 11.84LVIIGSS1324 pKa = 3.66
MM1 pKa = 7.38RR2 pKa = 11.84TLTTFLFFLAALNFIAGQSFRR23 pKa = 11.84TTLVSPATVSRR34 pKa = 11.84QTSNSDD40 pKa = 3.45LNGDD44 pKa = 3.94GHH46 pKa = 7.0QDD48 pKa = 4.08LITVNGEE55 pKa = 4.0VGWLKK60 pKa = 10.32WVVGAHH66 pKa = 7.41AYY68 pKa = 9.6PEE70 pKa = 4.14YY71 pKa = 10.89RR72 pKa = 11.84KK73 pKa = 8.39ITPDD77 pKa = 2.99EE78 pKa = 4.58NYY80 pKa = 8.93THH82 pKa = 7.45ALTGDD87 pKa = 3.33VDD89 pKa = 4.15SDD91 pKa = 3.77GDD93 pKa = 3.6IDD95 pKa = 4.92IVALGDD101 pKa = 3.73DD102 pKa = 3.63RR103 pKa = 11.84VVLLKK108 pKa = 11.28NNDD111 pKa = 3.28GEE113 pKa = 4.36AGFWLEE119 pKa = 5.07SMIDD123 pKa = 3.72DD124 pKa = 4.9EE125 pKa = 5.35EE126 pKa = 5.16VNDD129 pKa = 4.85PDD131 pKa = 3.5WGALGDD137 pKa = 3.74IDD139 pKa = 5.83GDD141 pKa = 3.91GDD143 pKa = 4.68LDD145 pKa = 4.24LLVASRR151 pKa = 11.84YY152 pKa = 9.87GYY154 pKa = 10.31YY155 pKa = 10.41LGWYY159 pKa = 7.79EE160 pKa = 4.11NTDD163 pKa = 3.24GTGEE167 pKa = 4.0FQFYY171 pKa = 11.32AEE173 pKa = 5.63LPDD176 pKa = 3.88AQNARR181 pKa = 11.84SFGALVDD188 pKa = 4.38FDD190 pKa = 4.04QDD192 pKa = 2.76GDD194 pKa = 4.45LDD196 pKa = 4.04VLSAGCYY203 pKa = 9.8LDD205 pKa = 5.5CTLEE209 pKa = 3.92WYY211 pKa = 9.0EE212 pKa = 3.99NLDD215 pKa = 3.55GEE217 pKa = 5.01GEE219 pKa = 3.95FGIRR223 pKa = 11.84KK224 pKa = 9.39EE225 pKa = 4.7IYY227 pKa = 9.23FDD229 pKa = 4.33DD230 pKa = 4.25YY231 pKa = 11.48GIEE234 pKa = 4.02NMVIDD239 pKa = 4.31VGDD242 pKa = 3.56VDD244 pKa = 3.89GDD246 pKa = 4.0GYY248 pKa = 11.02EE249 pKa = 5.63DD250 pKa = 4.12IMTTSTYY257 pKa = 10.17YY258 pKa = 10.7KK259 pKa = 9.6VIRR262 pKa = 11.84WHH264 pKa = 6.71RR265 pKa = 11.84NLIGSGEE272 pKa = 4.17MGFAAAVDD280 pKa = 3.36ISDD283 pKa = 4.19FDD285 pKa = 3.62WARR288 pKa = 11.84SIEE291 pKa = 4.38LTDD294 pKa = 4.74LDD296 pKa = 4.61NDD298 pKa = 3.48NDD300 pKa = 3.96LDD302 pKa = 3.81IAVAGDD308 pKa = 4.5DD309 pKa = 3.89DD310 pKa = 3.9THH312 pKa = 8.55LIGWYY317 pKa = 9.21EE318 pKa = 3.92NWSGDD323 pKa = 4.0GLFSNLNIIDD333 pKa = 4.78DD334 pKa = 4.31EE335 pKa = 4.49TDD337 pKa = 2.62VDD339 pKa = 4.29YY340 pKa = 11.71SRR342 pKa = 11.84LICFDD347 pKa = 2.99IDD349 pKa = 3.69EE350 pKa = 5.36NGTQDD355 pKa = 3.31ILTAGTLNSHH365 pKa = 6.57TIGWYY370 pKa = 10.37ANEE373 pKa = 4.25EE374 pKa = 4.18NTGSFGRR381 pKa = 11.84VNEE384 pKa = 4.1LVHH387 pKa = 6.75PYY389 pKa = 10.04IADD392 pKa = 4.05EE393 pKa = 4.25IMLNDD398 pKa = 3.28VDD400 pKa = 5.41GDD402 pKa = 4.17GNTDD406 pKa = 2.95IVAGGSWYY414 pKa = 9.75TNSGYY419 pKa = 10.93DD420 pKa = 3.33YY421 pKa = 11.09LAFSNIRR428 pKa = 11.84MVDD431 pKa = 3.66EE432 pKa = 4.91EE433 pKa = 4.55GHH435 pKa = 6.29LSLVEE440 pKa = 4.9DD441 pKa = 4.44FTNDD445 pKa = 3.94GITDD449 pKa = 5.94LITLEE454 pKa = 4.16RR455 pKa = 11.84LPLSTHH461 pKa = 5.78KK462 pKa = 10.79KK463 pKa = 7.45IVLYY467 pKa = 10.17EE468 pKa = 3.84RR469 pKa = 11.84SGEE472 pKa = 4.0QFIFSSDD479 pKa = 2.77IVEE482 pKa = 4.15VDD484 pKa = 2.97NASIKK489 pKa = 10.88NLVSLDD495 pKa = 3.56YY496 pKa = 11.56DD497 pKa = 3.56NDD499 pKa = 3.75GDD501 pKa = 5.17LDD503 pKa = 3.96FVYY506 pKa = 9.05QTAYY510 pKa = 10.8DD511 pKa = 4.14VVWLCEE517 pKa = 4.0NLGSDD522 pKa = 3.77NGFAAPVEE530 pKa = 4.24IADD533 pKa = 4.24TEE535 pKa = 4.38VSVNNFEE542 pKa = 5.91LGDD545 pKa = 4.1LDD547 pKa = 3.78GDD549 pKa = 4.31GYY551 pKa = 11.74VDD553 pKa = 4.43ILIGTDD559 pKa = 2.53WNQRR563 pKa = 11.84MGWYY567 pKa = 9.96RR568 pKa = 11.84NEE570 pKa = 4.27AGSGTFSDD578 pKa = 4.39LAIISSGQPYY588 pKa = 9.68EE589 pKa = 3.95QLDD592 pKa = 3.51WDD594 pKa = 4.49DD595 pKa = 3.77QDD597 pKa = 5.26GDD599 pKa = 3.78GDD601 pKa = 4.46LDD603 pKa = 3.75LVYY606 pKa = 10.71VYY608 pKa = 10.6EE609 pKa = 4.19EE610 pKa = 4.28NILRR614 pKa = 11.84RR615 pKa = 11.84NNEE618 pKa = 3.7GNGVFSSEE626 pKa = 3.79EE627 pKa = 4.01VVYY630 pKa = 10.63HH631 pKa = 6.84DD632 pKa = 4.19PAGGHH637 pKa = 4.9SFRR640 pKa = 11.84RR641 pKa = 11.84IYY643 pKa = 11.16CNDD646 pKa = 3.35HH647 pKa = 6.74NDD649 pKa = 3.43DD650 pKa = 3.82GLRR653 pKa = 11.84DD654 pKa = 3.83FLFFDD659 pKa = 3.36QSSYY663 pKa = 9.4GWRR666 pKa = 11.84FLTAEE671 pKa = 3.97IEE673 pKa = 4.08GGYY676 pKa = 10.12LLSAEE681 pKa = 4.6AEE683 pKa = 4.52FIPSNNVEE691 pKa = 4.37PIKK694 pKa = 10.72FYY696 pKa = 11.3DD697 pKa = 4.24LDD699 pKa = 4.09NDD701 pKa = 4.32GDD703 pKa = 4.43DD704 pKa = 5.75DD705 pKa = 4.09MLFAQGDD712 pKa = 3.8KK713 pKa = 11.03LYY715 pKa = 11.17VSEE718 pKa = 4.65NLNSGKK724 pKa = 9.89VLQGLIYY731 pKa = 10.51YY732 pKa = 9.99DD733 pKa = 3.86EE734 pKa = 4.77NEE736 pKa = 4.11NGINDD741 pKa = 4.35DD742 pKa = 3.78SDD744 pKa = 4.24SPLVDD749 pKa = 3.17QLVSIDD755 pKa = 3.53PTQLFLHH762 pKa = 6.72SNTDD766 pKa = 3.5GLFSTVLGNGQYY778 pKa = 9.22EE779 pKa = 4.58VRR781 pKa = 11.84CDD783 pKa = 4.38PEE785 pKa = 5.13DD786 pKa = 3.16GWGFVGDD793 pKa = 4.54SIVNIQTSSDD803 pKa = 3.28ADD805 pKa = 4.31TIWVEE810 pKa = 4.24FPLINTDD817 pKa = 3.17TTSRR821 pKa = 11.84PSVEE825 pKa = 4.26VTSGPVRR832 pKa = 11.84CGFEE836 pKa = 4.09VPFWLTVNNDD846 pKa = 2.99GYY848 pKa = 11.59VPEE851 pKa = 4.57TGDD854 pKa = 3.34FYY856 pKa = 11.33FYY858 pKa = 10.97LDD860 pKa = 3.99SLVALAIDD868 pKa = 4.42PADD871 pKa = 3.95HH872 pKa = 7.18PNLFVDD878 pKa = 3.82AEE880 pKa = 4.31GRR882 pKa = 11.84TGLFWEE888 pKa = 5.12QIDD891 pKa = 3.78ISPGARR897 pKa = 11.84WQTQVLLQMPSVDD910 pKa = 5.05FIGEE914 pKa = 3.84ALGFNGIFEE923 pKa = 4.49YY924 pKa = 10.86AVGINEE930 pKa = 4.03LQSNGEE936 pKa = 3.88YY937 pKa = 10.27TRR939 pKa = 11.84VLNCSYY945 pKa = 11.14DD946 pKa = 3.75PNDD949 pKa = 3.54KK950 pKa = 10.75SVEE953 pKa = 4.16EE954 pKa = 4.42NLPGYY959 pKa = 9.16PGYY962 pKa = 11.73SMVGDD967 pKa = 3.65SLEE970 pKa = 3.89YY971 pKa = 10.62LIRR974 pKa = 11.84FQNLGSDD981 pKa = 3.33TAINIKK987 pKa = 10.46VSDD990 pKa = 3.8QLSEE994 pKa = 3.95HH995 pKa = 6.74FDD997 pKa = 3.39RR998 pKa = 11.84SKK1000 pKa = 11.32LRR1002 pKa = 11.84ILSASHH1008 pKa = 6.94NYY1010 pKa = 10.07RR1011 pKa = 11.84MVLTDD1016 pKa = 4.08DD1017 pKa = 3.66NRR1019 pKa = 11.84LNVYY1023 pKa = 9.93FDD1025 pKa = 4.5NIYY1028 pKa = 10.86LPDD1031 pKa = 3.99AMTDD1035 pKa = 3.29EE1036 pKa = 5.05AGSHH1040 pKa = 5.74GFIKK1044 pKa = 10.39FRR1046 pKa = 11.84VGMLPGSTDD1055 pKa = 2.85GDD1057 pKa = 3.96VIEE1060 pKa = 4.8NQADD1064 pKa = 3.42IYY1066 pKa = 11.19FDD1068 pKa = 4.28FNPPITTNVVYY1079 pKa = 8.95NTILEE1084 pKa = 4.14RR1085 pKa = 11.84LIYY1088 pKa = 8.85PAQILAPKK1096 pKa = 9.98CFEE1099 pKa = 4.35FSDD1102 pKa = 4.25GSISVEE1108 pKa = 3.81EE1109 pKa = 4.21VVNGVDD1115 pKa = 3.84YY1116 pKa = 10.13FWNGISSSNHH1126 pKa = 5.54IEE1128 pKa = 4.02NLSAGTNTLEE1138 pKa = 4.7VYY1140 pKa = 10.56RR1141 pKa = 11.84SDD1143 pKa = 4.07GKK1145 pKa = 10.7ILADD1149 pKa = 3.4TSYY1152 pKa = 11.14QITDD1156 pKa = 3.58PDD1158 pKa = 3.64ILEE1161 pKa = 4.06VTAEE1165 pKa = 3.97IEE1167 pKa = 4.05PASFANSGSIIATPTGGTAPYY1188 pKa = 9.4TYY1190 pKa = 10.29VWQQFPEE1197 pKa = 4.07QQGNEE1202 pKa = 4.09IGDD1205 pKa = 4.08LAAGNYY1211 pKa = 9.09DD1212 pKa = 4.01LQVIDD1217 pKa = 5.18ANGCIIDD1224 pKa = 3.86AVFIVPEE1231 pKa = 4.21VVSTAEE1237 pKa = 4.25HH1238 pKa = 6.47LAKK1241 pKa = 10.66SPLSLRR1247 pKa = 11.84IYY1249 pKa = 9.3PNPARR1254 pKa = 11.84ALEE1257 pKa = 4.05FVNIRR1262 pKa = 11.84FSEE1265 pKa = 4.29VKK1267 pKa = 10.55NEE1269 pKa = 4.07SEE1271 pKa = 4.38LLVYY1275 pKa = 10.83DD1276 pKa = 3.47NLKK1279 pKa = 10.2RR1280 pKa = 11.84IVVRR1284 pKa = 11.84RR1285 pKa = 11.84SIAPGSYY1292 pKa = 10.29EE1293 pKa = 4.13LTDD1296 pKa = 4.11LKK1298 pKa = 10.69LAPGNYY1304 pKa = 9.27FLVLISGGKK1313 pKa = 8.11HH1314 pKa = 4.32ALGRR1318 pKa = 11.84LVIIGSS1324 pKa = 3.66
Molecular weight: 147.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C6BUT0|A0A2C6BUT0_9BACT Uncharacterized protein OS=Lewinellaceae bacterium SD302 OX=2014804 GN=CEQ90_19755 PE=4 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84HH3 pKa = 5.52RR4 pKa = 11.84QLPADD9 pKa = 4.01TSAASVTYY17 pKa = 9.85RR18 pKa = 11.84RR19 pKa = 11.84HH20 pKa = 4.45QFRR23 pKa = 11.84IFDD26 pKa = 4.01PSRR29 pKa = 11.84LKK31 pKa = 10.57IGSCVGAPVLYY42 pKa = 7.97NTPGKK47 pKa = 10.23NGQLPISEE55 pKa = 4.23IAIMILL61 pKa = 3.33
MM1 pKa = 8.02RR2 pKa = 11.84HH3 pKa = 5.52RR4 pKa = 11.84QLPADD9 pKa = 4.01TSAASVTYY17 pKa = 9.85RR18 pKa = 11.84RR19 pKa = 11.84HH20 pKa = 4.45QFRR23 pKa = 11.84IFDD26 pKa = 4.01PSRR29 pKa = 11.84LKK31 pKa = 10.57IGSCVGAPVLYY42 pKa = 7.97NTPGKK47 pKa = 10.23NGQLPISEE55 pKa = 4.23IAIMILL61 pKa = 3.33
Molecular weight: 6.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1596819 |
38 |
4969 |
394.1 |
43.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.856 ± 0.042 | 0.985 ± 0.027 |
6.346 ± 0.045 | 6.531 ± 0.039 |
4.713 ± 0.029 | 7.698 ± 0.037 |
1.717 ± 0.022 | 6.041 ± 0.03 |
4.233 ± 0.053 | 10.026 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.027 ± 0.02 | 4.875 ± 0.038 |
4.42 ± 0.03 | 3.661 ± 0.023 |
5.235 ± 0.05 | 6.207 ± 0.03 |
5.904 ± 0.063 | 6.545 ± 0.031 |
1.221 ± 0.017 | 3.758 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
