
Alistipes communis
Taxonomy: cellular organisms; Bacteria; FCB group;
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
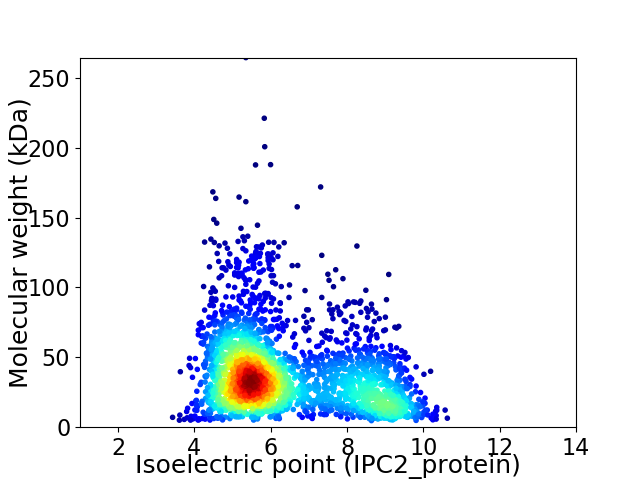
Virtual 2D-PAGE plot for 2673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1WQD6|A0A4Y1WQD6_9BACT ATPase OS=Alistipes communis OX=2585118 GN=A5CBH24_00470 PE=4 SV=1
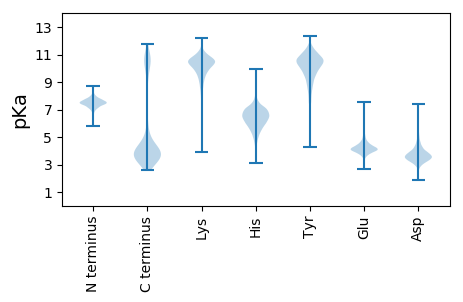
MM1 pKa = 7.46LLVALTGMGLASCSEE16 pKa = 4.27SPEE19 pKa = 4.05APEE22 pKa = 5.82DD23 pKa = 3.66PTEE26 pKa = 4.53NPDD29 pKa = 5.55DD30 pKa = 4.09EE31 pKa = 5.66DD32 pKa = 5.18KK33 pKa = 11.75GDD35 pKa = 3.62GSEE38 pKa = 5.04GEE40 pKa = 4.56GGDD43 pKa = 3.55EE44 pKa = 4.3NKK46 pKa = 9.67TVEE49 pKa = 4.23LNYY52 pKa = 10.01TVAEE56 pKa = 3.91AAYY59 pKa = 9.14WGDD62 pKa = 4.69DD63 pKa = 3.46YY64 pKa = 11.76MEE66 pKa = 6.13DD67 pKa = 3.55GTSNLIVYY75 pKa = 10.01LYY77 pKa = 10.1DD78 pKa = 3.45SPRR81 pKa = 11.84DD82 pKa = 3.33EE83 pKa = 5.86DD84 pKa = 4.3GVHH87 pKa = 6.63TGPKK91 pKa = 9.73DD92 pKa = 3.31HH93 pKa = 7.26FNIDD97 pKa = 3.22MNLPNFNGGEE107 pKa = 4.04LLIPARR113 pKa = 11.84SYY115 pKa = 10.94PIGEE119 pKa = 4.34PEE121 pKa = 4.28VYY123 pKa = 10.41APNTLEE129 pKa = 4.6PGFADD134 pKa = 3.48EE135 pKa = 5.09FEE137 pKa = 4.74IGGVLYY143 pKa = 10.84GDD145 pKa = 4.23YY146 pKa = 10.98YY147 pKa = 11.61GVFHH151 pKa = 6.04QVIDD155 pKa = 3.73AEE157 pKa = 4.31QNEE160 pKa = 4.73TYY162 pKa = 11.19EE163 pKa = 4.25MVSEE167 pKa = 4.58GEE169 pKa = 4.03MTVGYY174 pKa = 10.2EE175 pKa = 3.86NGVYY179 pKa = 10.19TIDD182 pKa = 3.3ATFTTEE188 pKa = 3.62EE189 pKa = 4.34GVVYY193 pKa = 10.38HH194 pKa = 5.5VTYY197 pKa = 10.51KK198 pKa = 11.01GAITVDD204 pKa = 4.08DD205 pKa = 4.57EE206 pKa = 5.17SLGEE210 pKa = 4.4PDD212 pKa = 5.32DD213 pKa = 4.39EE214 pKa = 4.68DD215 pKa = 3.77WYY217 pKa = 10.88STLTDD222 pKa = 3.9DD223 pKa = 5.38VEE225 pKa = 4.64VDD227 pKa = 4.17ALTQGQLYY235 pKa = 10.63YY236 pKa = 10.24MGADD240 pKa = 3.65DD241 pKa = 5.04KK242 pKa = 11.8SSLFGILLGTEE253 pKa = 4.53GVTLSNGTGTGEE265 pKa = 3.83LMQFFVNADD274 pKa = 3.31PDD276 pKa = 3.92AVKK279 pKa = 10.22EE280 pKa = 4.12LPAGKK285 pKa = 10.36YY286 pKa = 9.91EE287 pKa = 4.01IVPRR291 pKa = 11.84SAPMEE296 pKa = 4.45PEE298 pKa = 4.18TVIEE302 pKa = 5.0GYY304 pKa = 10.37KK305 pKa = 10.34EE306 pKa = 4.16GNQLGGSWYY315 pKa = 10.67FEE317 pKa = 4.18LEE319 pKa = 3.92KK320 pKa = 11.15NLTVNRR326 pKa = 11.84APAYY330 pKa = 9.23EE331 pKa = 4.07GTVTIGHH338 pKa = 5.96TEE340 pKa = 3.71NTLDD344 pKa = 3.51YY345 pKa = 10.23TVDD348 pKa = 3.89FEE350 pKa = 6.05FYY352 pKa = 10.81DD353 pKa = 4.09DD354 pKa = 5.31NLDD357 pKa = 3.12EE358 pKa = 5.16GYY360 pKa = 9.9IVSGSFSGKK369 pKa = 8.59MEE371 pKa = 4.5YY372 pKa = 9.85IYY374 pKa = 10.96DD375 pKa = 4.24AGTLQAKK382 pKa = 9.54GGRR385 pKa = 11.84VTAPRR390 pKa = 11.84RR391 pKa = 11.84PADD394 pKa = 3.24KK395 pKa = 9.48RR396 pKa = 11.84AARR399 pKa = 11.84IRR401 pKa = 11.84RR402 pKa = 3.65
MM1 pKa = 7.46LLVALTGMGLASCSEE16 pKa = 4.27SPEE19 pKa = 4.05APEE22 pKa = 5.82DD23 pKa = 3.66PTEE26 pKa = 4.53NPDD29 pKa = 5.55DD30 pKa = 4.09EE31 pKa = 5.66DD32 pKa = 5.18KK33 pKa = 11.75GDD35 pKa = 3.62GSEE38 pKa = 5.04GEE40 pKa = 4.56GGDD43 pKa = 3.55EE44 pKa = 4.3NKK46 pKa = 9.67TVEE49 pKa = 4.23LNYY52 pKa = 10.01TVAEE56 pKa = 3.91AAYY59 pKa = 9.14WGDD62 pKa = 4.69DD63 pKa = 3.46YY64 pKa = 11.76MEE66 pKa = 6.13DD67 pKa = 3.55GTSNLIVYY75 pKa = 10.01LYY77 pKa = 10.1DD78 pKa = 3.45SPRR81 pKa = 11.84DD82 pKa = 3.33EE83 pKa = 5.86DD84 pKa = 4.3GVHH87 pKa = 6.63TGPKK91 pKa = 9.73DD92 pKa = 3.31HH93 pKa = 7.26FNIDD97 pKa = 3.22MNLPNFNGGEE107 pKa = 4.04LLIPARR113 pKa = 11.84SYY115 pKa = 10.94PIGEE119 pKa = 4.34PEE121 pKa = 4.28VYY123 pKa = 10.41APNTLEE129 pKa = 4.6PGFADD134 pKa = 3.48EE135 pKa = 5.09FEE137 pKa = 4.74IGGVLYY143 pKa = 10.84GDD145 pKa = 4.23YY146 pKa = 10.98YY147 pKa = 11.61GVFHH151 pKa = 6.04QVIDD155 pKa = 3.73AEE157 pKa = 4.31QNEE160 pKa = 4.73TYY162 pKa = 11.19EE163 pKa = 4.25MVSEE167 pKa = 4.58GEE169 pKa = 4.03MTVGYY174 pKa = 10.2EE175 pKa = 3.86NGVYY179 pKa = 10.19TIDD182 pKa = 3.3ATFTTEE188 pKa = 3.62EE189 pKa = 4.34GVVYY193 pKa = 10.38HH194 pKa = 5.5VTYY197 pKa = 10.51KK198 pKa = 11.01GAITVDD204 pKa = 4.08DD205 pKa = 4.57EE206 pKa = 5.17SLGEE210 pKa = 4.4PDD212 pKa = 5.32DD213 pKa = 4.39EE214 pKa = 4.68DD215 pKa = 3.77WYY217 pKa = 10.88STLTDD222 pKa = 3.9DD223 pKa = 5.38VEE225 pKa = 4.64VDD227 pKa = 4.17ALTQGQLYY235 pKa = 10.63YY236 pKa = 10.24MGADD240 pKa = 3.65DD241 pKa = 5.04KK242 pKa = 11.8SSLFGILLGTEE253 pKa = 4.53GVTLSNGTGTGEE265 pKa = 3.83LMQFFVNADD274 pKa = 3.31PDD276 pKa = 3.92AVKK279 pKa = 10.22EE280 pKa = 4.12LPAGKK285 pKa = 10.36YY286 pKa = 9.91EE287 pKa = 4.01IVPRR291 pKa = 11.84SAPMEE296 pKa = 4.45PEE298 pKa = 4.18TVIEE302 pKa = 5.0GYY304 pKa = 10.37KK305 pKa = 10.34EE306 pKa = 4.16GNQLGGSWYY315 pKa = 10.67FEE317 pKa = 4.18LEE319 pKa = 3.92KK320 pKa = 11.15NLTVNRR326 pKa = 11.84APAYY330 pKa = 9.23EE331 pKa = 4.07GTVTIGHH338 pKa = 5.96TEE340 pKa = 3.71NTLDD344 pKa = 3.51YY345 pKa = 10.23TVDD348 pKa = 3.89FEE350 pKa = 6.05FYY352 pKa = 10.81DD353 pKa = 4.09DD354 pKa = 5.31NLDD357 pKa = 3.12EE358 pKa = 5.16GYY360 pKa = 9.9IVSGSFSGKK369 pKa = 8.59MEE371 pKa = 4.5YY372 pKa = 9.85IYY374 pKa = 10.96DD375 pKa = 4.24AGTLQAKK382 pKa = 9.54GGRR385 pKa = 11.84VTAPRR390 pKa = 11.84RR391 pKa = 11.84PADD394 pKa = 3.24KK395 pKa = 9.48RR396 pKa = 11.84AARR399 pKa = 11.84IRR401 pKa = 11.84RR402 pKa = 3.65
Molecular weight: 44.13 kDa
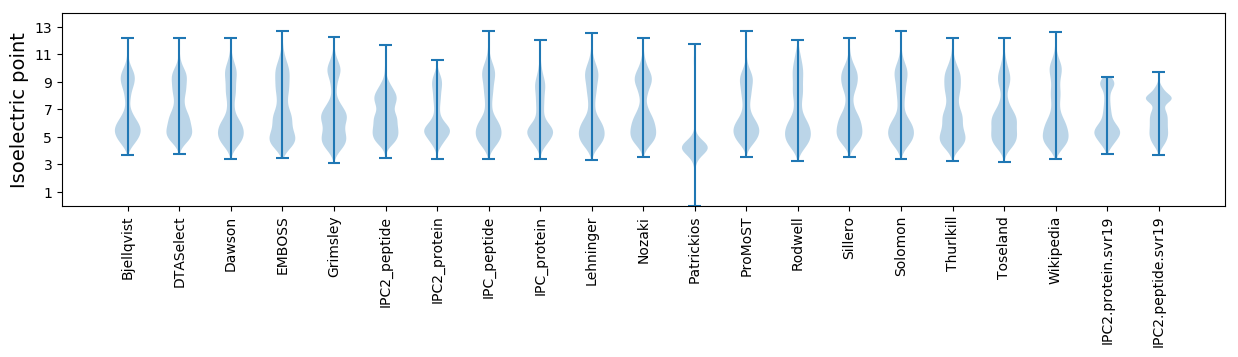
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1WUB2|A0A4Y1WUB2_9BACT Ribosome-recycling factor OS=Alistipes communis OX=2585118 GN=frr PE=3 SV=1
MM1 pKa = 6.74EE2 pKa = 5.23TKK4 pKa = 10.08LQHH7 pKa = 6.55PLLGEE12 pKa = 3.77IVLCQTRR19 pKa = 11.84RR20 pKa = 11.84ARR22 pKa = 11.84RR23 pKa = 11.84ISLSVRR29 pKa = 11.84STGGVRR35 pKa = 11.84LSFPCNVPRR44 pKa = 11.84ARR46 pKa = 11.84ALAFLEE52 pKa = 5.03SKK54 pKa = 9.3TDD56 pKa = 3.14WVIRR60 pKa = 11.84TRR62 pKa = 11.84DD63 pKa = 3.27RR64 pKa = 11.84LAARR68 pKa = 11.84AAAHH72 pKa = 6.58PAPAPCDD79 pKa = 3.37IEE81 pKa = 4.29ALRR84 pKa = 11.84SAAKK88 pKa = 10.49AEE90 pKa = 3.97LPTRR94 pKa = 11.84VEE96 pKa = 3.85EE97 pKa = 4.09LARR100 pKa = 11.84LHH102 pKa = 5.82GFRR105 pKa = 11.84YY106 pKa = 9.98GRR108 pKa = 11.84VTIRR112 pKa = 11.84ASRR115 pKa = 11.84TKK117 pKa = 9.64WGCCTGDD124 pKa = 3.46NNLSLSLFLMTLPPHH139 pKa = 6.55LRR141 pKa = 11.84DD142 pKa = 3.41FVLLHH147 pKa = 6.02EE148 pKa = 4.96LCHH151 pKa = 5.84TVHH154 pKa = 7.09HH155 pKa = 6.76DD156 pKa = 3.51HH157 pKa = 6.87SPHH160 pKa = 5.39FHH162 pKa = 6.92ALLDD166 pKa = 3.84RR167 pKa = 11.84LTGGNEE173 pKa = 3.69KK174 pKa = 10.54LLSRR178 pKa = 11.84EE179 pKa = 3.96LRR181 pKa = 11.84SYY183 pKa = 10.64SIPARR188 pKa = 3.79
MM1 pKa = 6.74EE2 pKa = 5.23TKK4 pKa = 10.08LQHH7 pKa = 6.55PLLGEE12 pKa = 3.77IVLCQTRR19 pKa = 11.84RR20 pKa = 11.84ARR22 pKa = 11.84RR23 pKa = 11.84ISLSVRR29 pKa = 11.84STGGVRR35 pKa = 11.84LSFPCNVPRR44 pKa = 11.84ARR46 pKa = 11.84ALAFLEE52 pKa = 5.03SKK54 pKa = 9.3TDD56 pKa = 3.14WVIRR60 pKa = 11.84TRR62 pKa = 11.84DD63 pKa = 3.27RR64 pKa = 11.84LAARR68 pKa = 11.84AAAHH72 pKa = 6.58PAPAPCDD79 pKa = 3.37IEE81 pKa = 4.29ALRR84 pKa = 11.84SAAKK88 pKa = 10.49AEE90 pKa = 3.97LPTRR94 pKa = 11.84VEE96 pKa = 3.85EE97 pKa = 4.09LARR100 pKa = 11.84LHH102 pKa = 5.82GFRR105 pKa = 11.84YY106 pKa = 9.98GRR108 pKa = 11.84VTIRR112 pKa = 11.84ASRR115 pKa = 11.84TKK117 pKa = 9.64WGCCTGDD124 pKa = 3.46NNLSLSLFLMTLPPHH139 pKa = 6.55LRR141 pKa = 11.84DD142 pKa = 3.41FVLLHH147 pKa = 6.02EE148 pKa = 4.96LCHH151 pKa = 5.84TVHH154 pKa = 7.09HH155 pKa = 6.76DD156 pKa = 3.51HH157 pKa = 6.87SPHH160 pKa = 5.39FHH162 pKa = 6.92ALLDD166 pKa = 3.84RR167 pKa = 11.84LTGGNEE173 pKa = 3.69KK174 pKa = 10.54LLSRR178 pKa = 11.84EE179 pKa = 3.96LRR181 pKa = 11.84SYY183 pKa = 10.64SIPARR188 pKa = 3.79
Molecular weight: 21.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
959588 |
41 |
2342 |
359.0 |
40.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.491 ± 0.064 | 1.311 ± 0.02 |
5.955 ± 0.034 | 6.559 ± 0.046 |
4.15 ± 0.025 | 7.54 ± 0.04 |
1.778 ± 0.017 | 5.546 ± 0.034 |
4.468 ± 0.045 | 9.038 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.394 ± 0.017 | 3.78 ± 0.031 |
4.117 ± 0.029 | 2.984 ± 0.024 |
6.845 ± 0.058 | 5.683 ± 0.044 |
5.848 ± 0.042 | 7.068 ± 0.037 |
1.282 ± 0.018 | 4.161 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
