
Dehalogenimonas alkenigignens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Dehalococcoidia; Dehalogenimonas
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
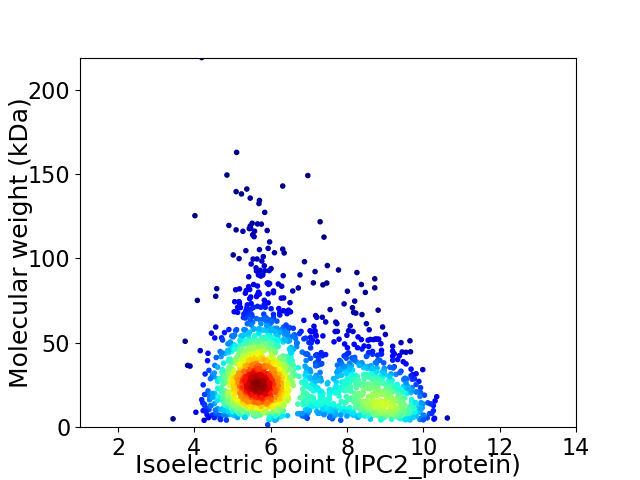
Virtual 2D-PAGE plot for 1927 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
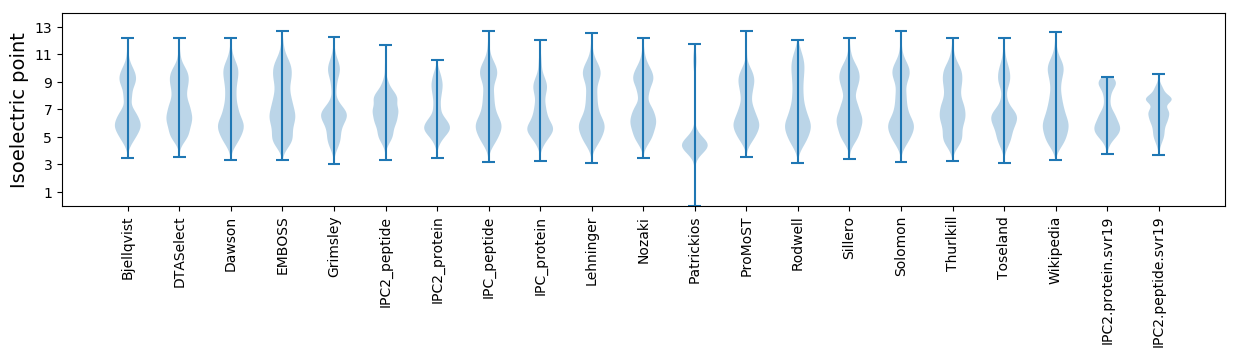
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W0GKE3|A0A0W0GKE3_9CHLR Uncharacterized protein OS=Dehalogenimonas alkenigignens OX=1217799 GN=DEALK_18410 PE=4 SV=1
MM1 pKa = 7.23KK2 pKa = 10.3RR3 pKa = 11.84VFKK6 pKa = 10.77KK7 pKa = 9.88FLVFAGVLAAVAAVLVPALAVPVSAQATNFSLNGYY42 pKa = 8.75RR43 pKa = 11.84VTVGGGGAYY52 pKa = 9.97SAGWTGGNLGNQWAEE67 pKa = 4.41GEE69 pKa = 4.3WVPYY73 pKa = 10.3KK74 pKa = 10.82LVITNVQTNYY84 pKa = 10.13PGLANFPDD92 pKa = 3.71IVLSFDD98 pKa = 3.27FHH100 pKa = 7.11DD101 pKa = 4.33ANGMFVDD108 pKa = 6.12LIRR111 pKa = 11.84GIQVGTTDD119 pKa = 3.62LTDD122 pKa = 3.63LQGFPTSGGGPQANNIAGAQLGQNDD147 pKa = 4.14PDD149 pKa = 3.01EE150 pKa = 4.43WAWTNFALLNLPNEE164 pKa = 4.29QVNRR168 pKa = 11.84SLVPLGDD175 pKa = 4.29LDD177 pKa = 4.2VAPGTDD183 pKa = 2.49KK184 pKa = 11.01HH185 pKa = 6.61VFIIHH190 pKa = 7.03PSDD193 pKa = 3.12ILGKK197 pKa = 10.79GGVTVDD203 pKa = 3.14SNTIVIYY210 pKa = 9.96YY211 pKa = 8.76QAHH214 pKa = 6.78LSRR217 pKa = 11.84TFVWSNALQDD227 pKa = 5.09FYY229 pKa = 11.76ATDD232 pKa = 3.66ADD234 pKa = 4.07LAKK237 pKa = 10.25WGGYY241 pKa = 9.96LYY243 pKa = 10.22ATDD246 pKa = 3.74AWPTSVQNGSGYY258 pKa = 9.93QPGSSGHH265 pKa = 4.92MHH267 pKa = 6.98LEE269 pKa = 4.15VEE271 pKa = 4.66GTGQQDD277 pKa = 3.63VPIPIPPKK285 pKa = 9.89PVGLIDD291 pKa = 4.26GYY293 pKa = 11.3KK294 pKa = 10.33FEE296 pKa = 6.24DD297 pKa = 3.99RR298 pKa = 11.84DD299 pKa = 4.22ADD301 pKa = 4.49HH302 pKa = 7.04IWDD305 pKa = 4.48PGEE308 pKa = 3.82PAIPGWDD315 pKa = 2.52IHH317 pKa = 6.32MYY319 pKa = 10.28SVLEE323 pKa = 3.95GGIVIDD329 pKa = 3.69VHH331 pKa = 5.65EE332 pKa = 4.53TTDD335 pKa = 3.42ANGFYY340 pKa = 10.46SFPNLTEE347 pKa = 4.51GIWYY351 pKa = 9.39IGEE354 pKa = 4.22HH355 pKa = 5.98VGPGDD360 pKa = 3.75PPLAGWTQTFPYY372 pKa = 10.27NVGNVPPGANSSLIDD387 pKa = 3.69NFPNNIKK394 pKa = 10.5AAIGPAHH401 pKa = 6.29LAEE404 pKa = 4.55WGWIVSLTDD413 pKa = 3.66AAPMQHH419 pKa = 5.79NVNFGNVSFATKK431 pKa = 9.85TGTKK435 pKa = 10.12FHH437 pKa = 7.4DD438 pKa = 4.07LNANGVWDD446 pKa = 3.86NGEE449 pKa = 4.31PGLGGWTIYY458 pKa = 10.94VDD460 pKa = 3.72YY461 pKa = 10.89NNNGVFDD468 pKa = 4.42AATEE472 pKa = 4.13PSAVTAGDD480 pKa = 3.42GTYY483 pKa = 10.35SINNVAPGTWRR494 pKa = 11.84VRR496 pKa = 11.84EE497 pKa = 3.95VLQAGWFASYY507 pKa = 9.22PATSDD512 pKa = 2.78AFGRR516 pKa = 11.84YY517 pKa = 8.92HH518 pKa = 7.38EE519 pKa = 4.33EE520 pKa = 4.12TFVGGQTYY528 pKa = 10.55SGNDD532 pKa = 3.2FGNWTTASKK541 pKa = 10.97SGMKK545 pKa = 10.27FEE547 pKa = 5.91DD548 pKa = 3.6MNADD552 pKa = 3.42GDD554 pKa = 4.16KK555 pKa = 11.0DD556 pKa = 4.4AEE558 pKa = 4.35DD559 pKa = 3.45TGLAGWVIYY568 pKa = 10.79VDD570 pKa = 3.95YY571 pKa = 10.88NDD573 pKa = 5.36NGVKK577 pKa = 10.22DD578 pKa = 3.97ANEE581 pKa = 4.1PFATTAADD589 pKa = 3.53GTYY592 pKa = 9.62TITGINPGTWKK603 pKa = 10.5VKK605 pKa = 10.18EE606 pKa = 4.02VAQAGWTQSYY616 pKa = 9.25PVSGYY621 pKa = 10.38HH622 pKa = 6.58EE623 pKa = 4.31EE624 pKa = 4.46TFTSGAALTGNDD636 pKa = 3.26FGNWTTAEE644 pKa = 3.82KK645 pKa = 10.46SGYY648 pKa = 9.59KK649 pKa = 10.54FNDD652 pKa = 3.45KK653 pKa = 10.75YY654 pKa = 11.42DD655 pKa = 3.98ADD657 pKa = 4.16GVRR660 pKa = 11.84DD661 pKa = 3.71ADD663 pKa = 3.86GVDD666 pKa = 3.82NILGNADD673 pKa = 4.3DD674 pKa = 4.75EE675 pKa = 4.8VLLSGWTIRR684 pKa = 11.84IYY686 pKa = 11.13EE687 pKa = 4.12DD688 pKa = 3.47TNGNGLLDD696 pKa = 3.66QAEE699 pKa = 4.37YY700 pKa = 10.96DD701 pKa = 3.69AGPLFTDD708 pKa = 3.42VTDD711 pKa = 3.6VNGAYY716 pKa = 10.42SFTVNPGKK724 pKa = 10.88YY725 pKa = 9.02IVVEE729 pKa = 4.11VMQDD733 pKa = 3.16KK734 pKa = 11.07LGTPYY739 pKa = 10.21YY740 pKa = 10.04QSYY743 pKa = 8.71PATPVLAGGLNTGTEE758 pKa = 4.07LLGANGHH765 pKa = 6.27AVTLVSSQIDD775 pKa = 3.86NNNHH779 pKa = 6.22FGNYY783 pKa = 7.39WPHH786 pKa = 4.62TTMTLFDD793 pKa = 4.67FVWEE797 pKa = 4.46TTADD801 pKa = 3.73GNVTLTISDD810 pKa = 3.99TNDD813 pKa = 2.87GDD815 pKa = 4.36VNLTDD820 pKa = 3.5AHH822 pKa = 5.78VHH824 pKa = 6.05LLANDD829 pKa = 3.37VEE831 pKa = 5.05YY832 pKa = 9.91PFSPISTPTSGDD844 pKa = 3.33DD845 pKa = 3.38GDD847 pKa = 5.67GIFEE851 pKa = 5.26PGEE854 pKa = 3.78TWTWIVYY861 pKa = 7.49VTITQDD867 pKa = 3.18TTFSTWGHH875 pKa = 6.02GEE877 pKa = 4.09DD878 pKa = 3.91PFGNTVDD885 pKa = 3.7YY886 pKa = 9.73PQYY889 pKa = 8.4QSEE892 pKa = 4.33YY893 pKa = 8.84AEE895 pKa = 4.1VFVEE899 pKa = 4.28VNEE902 pKa = 4.13ATRR905 pKa = 11.84TQGFWSTHH913 pKa = 4.8LDD915 pKa = 3.48FTEE918 pKa = 4.69MIFDD922 pKa = 4.3NYY924 pKa = 10.52TGDD927 pKa = 4.24EE928 pKa = 4.14VDD930 pKa = 3.5NDD932 pKa = 3.45NVGFIDD938 pKa = 4.63LGWRR942 pKa = 11.84QITNINDD949 pKa = 3.23LMGVFWANNAKK960 pKa = 10.58NSDD963 pKa = 3.27GSNRR967 pKa = 11.84SALCQARR974 pKa = 11.84IIASNQALAALLNAAMPGGAPLPAGYY1000 pKa = 10.19SPAEE1004 pKa = 3.64IAAILGGTDD1013 pKa = 2.81IPAIKK1018 pKa = 9.85MLGSVLGAYY1027 pKa = 10.21NEE1029 pKa = 5.04DD1030 pKa = 3.78GDD1032 pKa = 5.8DD1033 pKa = 3.75VALDD1037 pKa = 4.78PILGSTGKK1045 pKa = 10.5ANPQQAKK1052 pKa = 9.83ALANIGFADD1061 pKa = 4.31CNIQTSLVVAALPASVVKK1079 pKa = 8.78NTPVNITFTLNTVNDD1094 pKa = 3.42VGAAGGTFTVIASKK1108 pKa = 10.69DD1109 pKa = 3.55SKK1111 pKa = 10.99FPNGANKK1118 pKa = 9.64TITVGSGAVNDD1129 pKa = 3.83MPFNGGVSFTPTSAGTWYY1147 pKa = 10.13IKK1149 pKa = 9.12ITYY1152 pKa = 10.02SGDD1155 pKa = 3.1GEE1157 pKa = 4.65NYY1159 pKa = 9.52GGCSVIVTLVVTNSS1173 pKa = 2.91
MM1 pKa = 7.23KK2 pKa = 10.3RR3 pKa = 11.84VFKK6 pKa = 10.77KK7 pKa = 9.88FLVFAGVLAAVAAVLVPALAVPVSAQATNFSLNGYY42 pKa = 8.75RR43 pKa = 11.84VTVGGGGAYY52 pKa = 9.97SAGWTGGNLGNQWAEE67 pKa = 4.41GEE69 pKa = 4.3WVPYY73 pKa = 10.3KK74 pKa = 10.82LVITNVQTNYY84 pKa = 10.13PGLANFPDD92 pKa = 3.71IVLSFDD98 pKa = 3.27FHH100 pKa = 7.11DD101 pKa = 4.33ANGMFVDD108 pKa = 6.12LIRR111 pKa = 11.84GIQVGTTDD119 pKa = 3.62LTDD122 pKa = 3.63LQGFPTSGGGPQANNIAGAQLGQNDD147 pKa = 4.14PDD149 pKa = 3.01EE150 pKa = 4.43WAWTNFALLNLPNEE164 pKa = 4.29QVNRR168 pKa = 11.84SLVPLGDD175 pKa = 4.29LDD177 pKa = 4.2VAPGTDD183 pKa = 2.49KK184 pKa = 11.01HH185 pKa = 6.61VFIIHH190 pKa = 7.03PSDD193 pKa = 3.12ILGKK197 pKa = 10.79GGVTVDD203 pKa = 3.14SNTIVIYY210 pKa = 9.96YY211 pKa = 8.76QAHH214 pKa = 6.78LSRR217 pKa = 11.84TFVWSNALQDD227 pKa = 5.09FYY229 pKa = 11.76ATDD232 pKa = 3.66ADD234 pKa = 4.07LAKK237 pKa = 10.25WGGYY241 pKa = 9.96LYY243 pKa = 10.22ATDD246 pKa = 3.74AWPTSVQNGSGYY258 pKa = 9.93QPGSSGHH265 pKa = 4.92MHH267 pKa = 6.98LEE269 pKa = 4.15VEE271 pKa = 4.66GTGQQDD277 pKa = 3.63VPIPIPPKK285 pKa = 9.89PVGLIDD291 pKa = 4.26GYY293 pKa = 11.3KK294 pKa = 10.33FEE296 pKa = 6.24DD297 pKa = 3.99RR298 pKa = 11.84DD299 pKa = 4.22ADD301 pKa = 4.49HH302 pKa = 7.04IWDD305 pKa = 4.48PGEE308 pKa = 3.82PAIPGWDD315 pKa = 2.52IHH317 pKa = 6.32MYY319 pKa = 10.28SVLEE323 pKa = 3.95GGIVIDD329 pKa = 3.69VHH331 pKa = 5.65EE332 pKa = 4.53TTDD335 pKa = 3.42ANGFYY340 pKa = 10.46SFPNLTEE347 pKa = 4.51GIWYY351 pKa = 9.39IGEE354 pKa = 4.22HH355 pKa = 5.98VGPGDD360 pKa = 3.75PPLAGWTQTFPYY372 pKa = 10.27NVGNVPPGANSSLIDD387 pKa = 3.69NFPNNIKK394 pKa = 10.5AAIGPAHH401 pKa = 6.29LAEE404 pKa = 4.55WGWIVSLTDD413 pKa = 3.66AAPMQHH419 pKa = 5.79NVNFGNVSFATKK431 pKa = 9.85TGTKK435 pKa = 10.12FHH437 pKa = 7.4DD438 pKa = 4.07LNANGVWDD446 pKa = 3.86NGEE449 pKa = 4.31PGLGGWTIYY458 pKa = 10.94VDD460 pKa = 3.72YY461 pKa = 10.89NNNGVFDD468 pKa = 4.42AATEE472 pKa = 4.13PSAVTAGDD480 pKa = 3.42GTYY483 pKa = 10.35SINNVAPGTWRR494 pKa = 11.84VRR496 pKa = 11.84EE497 pKa = 3.95VLQAGWFASYY507 pKa = 9.22PATSDD512 pKa = 2.78AFGRR516 pKa = 11.84YY517 pKa = 8.92HH518 pKa = 7.38EE519 pKa = 4.33EE520 pKa = 4.12TFVGGQTYY528 pKa = 10.55SGNDD532 pKa = 3.2FGNWTTASKK541 pKa = 10.97SGMKK545 pKa = 10.27FEE547 pKa = 5.91DD548 pKa = 3.6MNADD552 pKa = 3.42GDD554 pKa = 4.16KK555 pKa = 11.0DD556 pKa = 4.4AEE558 pKa = 4.35DD559 pKa = 3.45TGLAGWVIYY568 pKa = 10.79VDD570 pKa = 3.95YY571 pKa = 10.88NDD573 pKa = 5.36NGVKK577 pKa = 10.22DD578 pKa = 3.97ANEE581 pKa = 4.1PFATTAADD589 pKa = 3.53GTYY592 pKa = 9.62TITGINPGTWKK603 pKa = 10.5VKK605 pKa = 10.18EE606 pKa = 4.02VAQAGWTQSYY616 pKa = 9.25PVSGYY621 pKa = 10.38HH622 pKa = 6.58EE623 pKa = 4.31EE624 pKa = 4.46TFTSGAALTGNDD636 pKa = 3.26FGNWTTAEE644 pKa = 3.82KK645 pKa = 10.46SGYY648 pKa = 9.59KK649 pKa = 10.54FNDD652 pKa = 3.45KK653 pKa = 10.75YY654 pKa = 11.42DD655 pKa = 3.98ADD657 pKa = 4.16GVRR660 pKa = 11.84DD661 pKa = 3.71ADD663 pKa = 3.86GVDD666 pKa = 3.82NILGNADD673 pKa = 4.3DD674 pKa = 4.75EE675 pKa = 4.8VLLSGWTIRR684 pKa = 11.84IYY686 pKa = 11.13EE687 pKa = 4.12DD688 pKa = 3.47TNGNGLLDD696 pKa = 3.66QAEE699 pKa = 4.37YY700 pKa = 10.96DD701 pKa = 3.69AGPLFTDD708 pKa = 3.42VTDD711 pKa = 3.6VNGAYY716 pKa = 10.42SFTVNPGKK724 pKa = 10.88YY725 pKa = 9.02IVVEE729 pKa = 4.11VMQDD733 pKa = 3.16KK734 pKa = 11.07LGTPYY739 pKa = 10.21YY740 pKa = 10.04QSYY743 pKa = 8.71PATPVLAGGLNTGTEE758 pKa = 4.07LLGANGHH765 pKa = 6.27AVTLVSSQIDD775 pKa = 3.86NNNHH779 pKa = 6.22FGNYY783 pKa = 7.39WPHH786 pKa = 4.62TTMTLFDD793 pKa = 4.67FVWEE797 pKa = 4.46TTADD801 pKa = 3.73GNVTLTISDD810 pKa = 3.99TNDD813 pKa = 2.87GDD815 pKa = 4.36VNLTDD820 pKa = 3.5AHH822 pKa = 5.78VHH824 pKa = 6.05LLANDD829 pKa = 3.37VEE831 pKa = 5.05YY832 pKa = 9.91PFSPISTPTSGDD844 pKa = 3.33DD845 pKa = 3.38GDD847 pKa = 5.67GIFEE851 pKa = 5.26PGEE854 pKa = 3.78TWTWIVYY861 pKa = 7.49VTITQDD867 pKa = 3.18TTFSTWGHH875 pKa = 6.02GEE877 pKa = 4.09DD878 pKa = 3.91PFGNTVDD885 pKa = 3.7YY886 pKa = 9.73PQYY889 pKa = 8.4QSEE892 pKa = 4.33YY893 pKa = 8.84AEE895 pKa = 4.1VFVEE899 pKa = 4.28VNEE902 pKa = 4.13ATRR905 pKa = 11.84TQGFWSTHH913 pKa = 4.8LDD915 pKa = 3.48FTEE918 pKa = 4.69MIFDD922 pKa = 4.3NYY924 pKa = 10.52TGDD927 pKa = 4.24EE928 pKa = 4.14VDD930 pKa = 3.5NDD932 pKa = 3.45NVGFIDD938 pKa = 4.63LGWRR942 pKa = 11.84QITNINDD949 pKa = 3.23LMGVFWANNAKK960 pKa = 10.58NSDD963 pKa = 3.27GSNRR967 pKa = 11.84SALCQARR974 pKa = 11.84IIASNQALAALLNAAMPGGAPLPAGYY1000 pKa = 10.19SPAEE1004 pKa = 3.64IAAILGGTDD1013 pKa = 2.81IPAIKK1018 pKa = 9.85MLGSVLGAYY1027 pKa = 10.21NEE1029 pKa = 5.04DD1030 pKa = 3.78GDD1032 pKa = 5.8DD1033 pKa = 3.75VALDD1037 pKa = 4.78PILGSTGKK1045 pKa = 10.5ANPQQAKK1052 pKa = 9.83ALANIGFADD1061 pKa = 4.31CNIQTSLVVAALPASVVKK1079 pKa = 8.78NTPVNITFTLNTVNDD1094 pKa = 3.42VGAAGGTFTVIASKK1108 pKa = 10.69DD1109 pKa = 3.55SKK1111 pKa = 10.99FPNGANKK1118 pKa = 9.64TITVGSGAVNDD1129 pKa = 3.83MPFNGGVSFTPTSAGTWYY1147 pKa = 10.13IKK1149 pKa = 9.12ITYY1152 pKa = 10.02SGDD1155 pKa = 3.1GEE1157 pKa = 4.65NYY1159 pKa = 9.52GGCSVIVTLVVTNSS1173 pKa = 2.91
Molecular weight: 125.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W0GG97|A0A0W0GG97_9CHLR Probable transaldolase OS=Dehalogenimonas alkenigignens OX=1217799 GN=tal PE=3 SV=1
MM1 pKa = 7.83PSLKK5 pKa = 10.7AFFTNWWAPMPLRR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.64IYY22 pKa = 10.32LAFRR26 pKa = 11.84NNWIKK31 pKa = 10.27ISRR34 pKa = 11.84RR35 pKa = 11.84QSCCGHH41 pKa = 6.75PGEE44 pKa = 5.14PGCC47 pKa = 5.73
MM1 pKa = 7.83PSLKK5 pKa = 10.7AFFTNWWAPMPLRR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.64IYY22 pKa = 10.32LAFRR26 pKa = 11.84NNWIKK31 pKa = 10.27ISRR34 pKa = 11.84RR35 pKa = 11.84QSCCGHH41 pKa = 6.75PGEE44 pKa = 5.14PGCC47 pKa = 5.73
Molecular weight: 5.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
551133 |
13 |
2090 |
286.0 |
31.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.235 ± 0.069 | 1.188 ± 0.028 |
5.089 ± 0.037 | 6.244 ± 0.062 |
3.936 ± 0.04 | 8.132 ± 0.049 |
1.864 ± 0.023 | 6.417 ± 0.051 |
4.791 ± 0.058 | 10.047 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.408 ± 0.024 | 3.186 ± 0.041 |
4.838 ± 0.043 | 3.022 ± 0.027 |
6.103 ± 0.057 | 5.925 ± 0.048 |
5.375 ± 0.065 | 7.298 ± 0.044 |
1.158 ± 0.022 | 2.744 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
