
Lake Sarah-associated circular virus-14
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
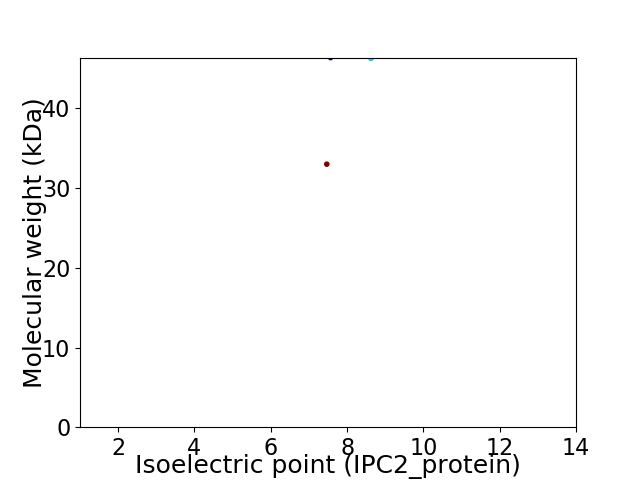
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
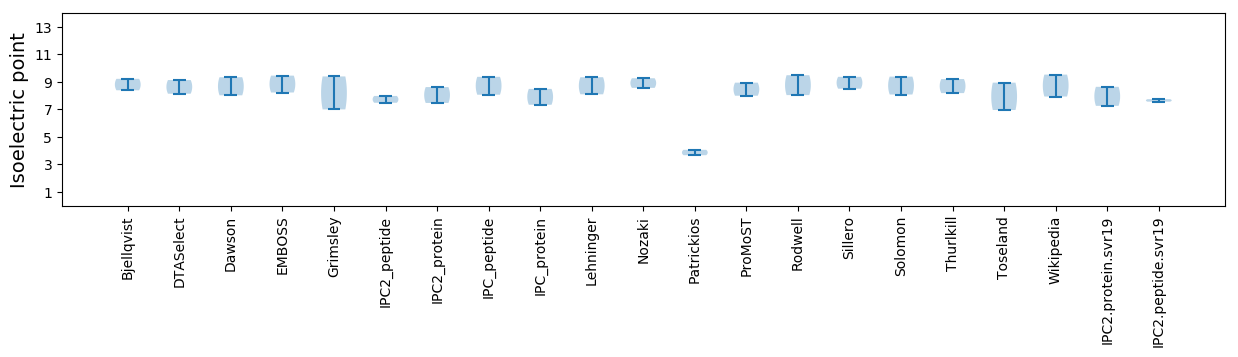
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA43|A0A126GA43_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-14 OX=1685740 PE=3 SV=1
MM1 pKa = 7.32PSSYY5 pKa = 10.36RR6 pKa = 11.84RR7 pKa = 11.84VVFTLNNYY15 pKa = 7.42TNEE18 pKa = 4.03ILTSICNYY26 pKa = 10.35AEE28 pKa = 4.34SNCRR32 pKa = 11.84YY33 pKa = 10.25AIIAKK38 pKa = 9.4EE39 pKa = 3.98VAPTTGTPHH48 pKa = 6.32LQGFLHH54 pKa = 6.57FKK56 pKa = 10.14NPKK59 pKa = 4.89THH61 pKa = 5.91KK62 pKa = 9.26TLGKK66 pKa = 9.57VLPGGHH72 pKa = 6.62FLHH75 pKa = 7.23AKK77 pKa = 10.44GSDD80 pKa = 3.44EE81 pKa = 4.79DD82 pKa = 4.05SQVYY86 pKa = 9.72CSKK89 pKa = 10.51EE90 pKa = 4.0DD91 pKa = 3.61PSPWEE96 pKa = 4.15FGTMCHH102 pKa = 5.63QGKK105 pKa = 10.26RR106 pKa = 11.84SDD108 pKa = 4.21LDD110 pKa = 3.84DD111 pKa = 6.27AILTLNEE118 pKa = 4.12SGGDD122 pKa = 3.63LKK124 pKa = 10.74RR125 pKa = 11.84VAMEE129 pKa = 3.56HH130 pKa = 5.93AGAYY134 pKa = 8.75VRR136 pKa = 11.84YY137 pKa = 8.97HH138 pKa = 6.85RR139 pKa = 11.84GFAAYY144 pKa = 9.29KK145 pKa = 10.44SLVCATAPRR154 pKa = 11.84DD155 pKa = 3.86FKK157 pKa = 10.99TKK159 pKa = 10.44LVVLFGPPGTGKK171 pKa = 8.03TRR173 pKa = 11.84AAYY176 pKa = 9.27EE177 pKa = 3.89LAGEE181 pKa = 4.22NPYY184 pKa = 10.43PKK186 pKa = 9.93PRR188 pKa = 11.84GEE190 pKa = 3.73WWDD193 pKa = 4.54GYY195 pKa = 10.68CGNNGVIIDD204 pKa = 5.2DD205 pKa = 4.44FYY207 pKa = 11.9GWLKK211 pKa = 10.53FDD213 pKa = 4.24EE214 pKa = 4.6LLKK217 pKa = 10.86ISDD220 pKa = 4.19RR221 pKa = 11.84YY222 pKa = 9.39PYY224 pKa = 10.07RR225 pKa = 11.84VPIKK229 pKa = 10.33GGYY232 pKa = 9.47EE233 pKa = 3.93NFCSKK238 pKa = 10.54IIIITSNIDD247 pKa = 2.61ISKK250 pKa = 8.93WYY252 pKa = 10.3KK253 pKa = 10.02FDD255 pKa = 4.38GYY257 pKa = 11.21DD258 pKa = 3.16PAALYY263 pKa = 9.93RR264 pKa = 11.84RR265 pKa = 11.84CTKK268 pKa = 10.08YY269 pKa = 10.51LRR271 pKa = 11.84CEE273 pKa = 3.93KK274 pKa = 10.79DD275 pKa = 3.54VFCSMEE281 pKa = 4.46DD282 pKa = 3.54DD283 pKa = 3.84YY284 pKa = 11.91LVKK287 pKa = 10.43INYY290 pKa = 8.84
MM1 pKa = 7.32PSSYY5 pKa = 10.36RR6 pKa = 11.84RR7 pKa = 11.84VVFTLNNYY15 pKa = 7.42TNEE18 pKa = 4.03ILTSICNYY26 pKa = 10.35AEE28 pKa = 4.34SNCRR32 pKa = 11.84YY33 pKa = 10.25AIIAKK38 pKa = 9.4EE39 pKa = 3.98VAPTTGTPHH48 pKa = 6.32LQGFLHH54 pKa = 6.57FKK56 pKa = 10.14NPKK59 pKa = 4.89THH61 pKa = 5.91KK62 pKa = 9.26TLGKK66 pKa = 9.57VLPGGHH72 pKa = 6.62FLHH75 pKa = 7.23AKK77 pKa = 10.44GSDD80 pKa = 3.44EE81 pKa = 4.79DD82 pKa = 4.05SQVYY86 pKa = 9.72CSKK89 pKa = 10.51EE90 pKa = 4.0DD91 pKa = 3.61PSPWEE96 pKa = 4.15FGTMCHH102 pKa = 5.63QGKK105 pKa = 10.26RR106 pKa = 11.84SDD108 pKa = 4.21LDD110 pKa = 3.84DD111 pKa = 6.27AILTLNEE118 pKa = 4.12SGGDD122 pKa = 3.63LKK124 pKa = 10.74RR125 pKa = 11.84VAMEE129 pKa = 3.56HH130 pKa = 5.93AGAYY134 pKa = 8.75VRR136 pKa = 11.84YY137 pKa = 8.97HH138 pKa = 6.85RR139 pKa = 11.84GFAAYY144 pKa = 9.29KK145 pKa = 10.44SLVCATAPRR154 pKa = 11.84DD155 pKa = 3.86FKK157 pKa = 10.99TKK159 pKa = 10.44LVVLFGPPGTGKK171 pKa = 8.03TRR173 pKa = 11.84AAYY176 pKa = 9.27EE177 pKa = 3.89LAGEE181 pKa = 4.22NPYY184 pKa = 10.43PKK186 pKa = 9.93PRR188 pKa = 11.84GEE190 pKa = 3.73WWDD193 pKa = 4.54GYY195 pKa = 10.68CGNNGVIIDD204 pKa = 5.2DD205 pKa = 4.44FYY207 pKa = 11.9GWLKK211 pKa = 10.53FDD213 pKa = 4.24EE214 pKa = 4.6LLKK217 pKa = 10.86ISDD220 pKa = 4.19RR221 pKa = 11.84YY222 pKa = 9.39PYY224 pKa = 10.07RR225 pKa = 11.84VPIKK229 pKa = 10.33GGYY232 pKa = 9.47EE233 pKa = 3.93NFCSKK238 pKa = 10.54IIIITSNIDD247 pKa = 2.61ISKK250 pKa = 8.93WYY252 pKa = 10.3KK253 pKa = 10.02FDD255 pKa = 4.38GYY257 pKa = 11.21DD258 pKa = 3.16PAALYY263 pKa = 9.93RR264 pKa = 11.84RR265 pKa = 11.84CTKK268 pKa = 10.08YY269 pKa = 10.51LRR271 pKa = 11.84CEE273 pKa = 3.93KK274 pKa = 10.79DD275 pKa = 3.54VFCSMEE281 pKa = 4.46DD282 pKa = 3.54DD283 pKa = 3.84YY284 pKa = 11.91LVKK287 pKa = 10.43INYY290 pKa = 8.84
Molecular weight: 33.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA43|A0A126GA43_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-14 OX=1685740 PE=3 SV=1
MM1 pKa = 7.76PRR3 pKa = 11.84TKK5 pKa = 9.96TYY7 pKa = 10.39RR8 pKa = 11.84KK9 pKa = 10.01KK10 pKa = 10.73SGKK13 pKa = 9.84KK14 pKa = 8.29YY15 pKa = 10.59NKK17 pKa = 9.27QKK19 pKa = 9.74TYY21 pKa = 10.38RR22 pKa = 11.84KK23 pKa = 9.69KK24 pKa = 10.9SYY26 pKa = 8.42GTKK29 pKa = 9.27KK30 pKa = 10.16KK31 pKa = 9.83SYY33 pKa = 10.11KK34 pKa = 8.88RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.68YY38 pKa = 10.4GNPVNQPMVSKK49 pKa = 10.71SLSAEE54 pKa = 3.67QAYY57 pKa = 10.9LKK59 pKa = 10.2MIMNPCEE66 pKa = 3.89SLLAYY71 pKa = 10.05GPSNGEE77 pKa = 3.72IEE79 pKa = 4.36SGYY82 pKa = 11.16LMRR85 pKa = 11.84TTYY88 pKa = 10.08RR89 pKa = 11.84VKK91 pKa = 10.27LHH93 pKa = 6.85RR94 pKa = 11.84IKK96 pKa = 10.84EE97 pKa = 4.11RR98 pKa = 11.84FNGYY102 pKa = 8.97VIWFPDD108 pKa = 3.5FTNAGIGSQVKK119 pKa = 10.31ANNPYY124 pKa = 10.43YY125 pKa = 10.32DD126 pKa = 4.16ANGTDD131 pKa = 3.76VKK133 pKa = 11.23NLTAGNCFVWEE144 pKa = 4.5CNDD147 pKa = 4.43AGAIPQNSTIGIAPATTSAYY167 pKa = 9.99YY168 pKa = 10.49GYY170 pKa = 10.86GFSDD174 pKa = 3.7LGDD177 pKa = 3.95GGSASSIKK185 pKa = 10.73DD186 pKa = 3.16PAYY189 pKa = 10.29NWASANIVSQIRR201 pKa = 11.84CLAACMTWQYY211 pKa = 11.29TGTLKK216 pKa = 10.12DD217 pKa = 3.59TAGQMCTVANITPYY231 pKa = 11.12QLMYY235 pKa = 11.34GNGGTDD241 pKa = 3.32NAPISISDD249 pKa = 3.96MISLNKK255 pKa = 10.37SYY257 pKa = 11.35DD258 pKa = 3.53RR259 pKa = 11.84IAVQKK264 pKa = 10.75YY265 pKa = 6.75EE266 pKa = 4.26VKK268 pKa = 9.2HH269 pKa = 5.46TPSTLGSLYY278 pKa = 10.42RR279 pKa = 11.84DD280 pKa = 3.25TDD282 pKa = 3.32KK283 pKa = 11.11PYY285 pKa = 10.39TDD287 pKa = 3.97AATGQTSKK295 pKa = 9.79TGDD298 pKa = 3.33TNGNCFVKK306 pKa = 10.66GNAVEE311 pKa = 4.33TVDD314 pKa = 3.67EE315 pKa = 4.78FGVGALTGTTPTTISLEE332 pKa = 3.93ASHH335 pKa = 6.93INGIGFCWTGIDD347 pKa = 3.36ATVAQNIEE355 pKa = 3.75IEE357 pKa = 4.16MTKK360 pKa = 9.93IFEE363 pKa = 4.23WTPSGKK369 pKa = 10.06YY370 pKa = 9.91LATSSNAVSRR380 pKa = 11.84PIMTTDD386 pKa = 3.65QIMSSLNNRR395 pKa = 11.84MPGWEE400 pKa = 4.14SRR402 pKa = 11.84HH403 pKa = 5.13QRR405 pKa = 11.84MTPAQVSAVSGQAGVPNGGG424 pKa = 3.31
MM1 pKa = 7.76PRR3 pKa = 11.84TKK5 pKa = 9.96TYY7 pKa = 10.39RR8 pKa = 11.84KK9 pKa = 10.01KK10 pKa = 10.73SGKK13 pKa = 9.84KK14 pKa = 8.29YY15 pKa = 10.59NKK17 pKa = 9.27QKK19 pKa = 9.74TYY21 pKa = 10.38RR22 pKa = 11.84KK23 pKa = 9.69KK24 pKa = 10.9SYY26 pKa = 8.42GTKK29 pKa = 9.27KK30 pKa = 10.16KK31 pKa = 9.83SYY33 pKa = 10.11KK34 pKa = 8.88RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.68YY38 pKa = 10.4GNPVNQPMVSKK49 pKa = 10.71SLSAEE54 pKa = 3.67QAYY57 pKa = 10.9LKK59 pKa = 10.2MIMNPCEE66 pKa = 3.89SLLAYY71 pKa = 10.05GPSNGEE77 pKa = 3.72IEE79 pKa = 4.36SGYY82 pKa = 11.16LMRR85 pKa = 11.84TTYY88 pKa = 10.08RR89 pKa = 11.84VKK91 pKa = 10.27LHH93 pKa = 6.85RR94 pKa = 11.84IKK96 pKa = 10.84EE97 pKa = 4.11RR98 pKa = 11.84FNGYY102 pKa = 8.97VIWFPDD108 pKa = 3.5FTNAGIGSQVKK119 pKa = 10.31ANNPYY124 pKa = 10.43YY125 pKa = 10.32DD126 pKa = 4.16ANGTDD131 pKa = 3.76VKK133 pKa = 11.23NLTAGNCFVWEE144 pKa = 4.5CNDD147 pKa = 4.43AGAIPQNSTIGIAPATTSAYY167 pKa = 9.99YY168 pKa = 10.49GYY170 pKa = 10.86GFSDD174 pKa = 3.7LGDD177 pKa = 3.95GGSASSIKK185 pKa = 10.73DD186 pKa = 3.16PAYY189 pKa = 10.29NWASANIVSQIRR201 pKa = 11.84CLAACMTWQYY211 pKa = 11.29TGTLKK216 pKa = 10.12DD217 pKa = 3.59TAGQMCTVANITPYY231 pKa = 11.12QLMYY235 pKa = 11.34GNGGTDD241 pKa = 3.32NAPISISDD249 pKa = 3.96MISLNKK255 pKa = 10.37SYY257 pKa = 11.35DD258 pKa = 3.53RR259 pKa = 11.84IAVQKK264 pKa = 10.75YY265 pKa = 6.75EE266 pKa = 4.26VKK268 pKa = 9.2HH269 pKa = 5.46TPSTLGSLYY278 pKa = 10.42RR279 pKa = 11.84DD280 pKa = 3.25TDD282 pKa = 3.32KK283 pKa = 11.11PYY285 pKa = 10.39TDD287 pKa = 3.97AATGQTSKK295 pKa = 9.79TGDD298 pKa = 3.33TNGNCFVKK306 pKa = 10.66GNAVEE311 pKa = 4.33TVDD314 pKa = 3.67EE315 pKa = 4.78FGVGALTGTTPTTISLEE332 pKa = 3.93ASHH335 pKa = 6.93INGIGFCWTGIDD347 pKa = 3.36ATVAQNIEE355 pKa = 3.75IEE357 pKa = 4.16MTKK360 pKa = 9.93IFEE363 pKa = 4.23WTPSGKK369 pKa = 10.06YY370 pKa = 9.91LATSSNAVSRR380 pKa = 11.84PIMTTDD386 pKa = 3.65QIMSSLNNRR395 pKa = 11.84MPGWEE400 pKa = 4.14SRR402 pKa = 11.84HH403 pKa = 5.13QRR405 pKa = 11.84MTPAQVSAVSGQAGVPNGGG424 pKa = 3.31
Molecular weight: 46.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
714 |
290 |
424 |
357.0 |
39.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.283 ± 0.636 | 2.521 ± 0.548 |
5.182 ± 0.809 | 4.062 ± 0.656 |
3.081 ± 0.828 | 8.964 ± 0.406 |
1.681 ± 0.637 | 5.742 ± 0.133 |
7.423 ± 0.3 | 5.462 ± 1.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.521 ± 0.674 | 5.882 ± 0.827 |
5.042 ± 0.281 | 2.661 ± 0.961 |
4.342 ± 0.491 | 7.283 ± 1.043 |
7.983 ± 1.457 | 4.762 ± 0.039 |
1.681 ± 0.026 | 6.443 ± 0.472 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
