
Hubei tombus-like virus 22
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
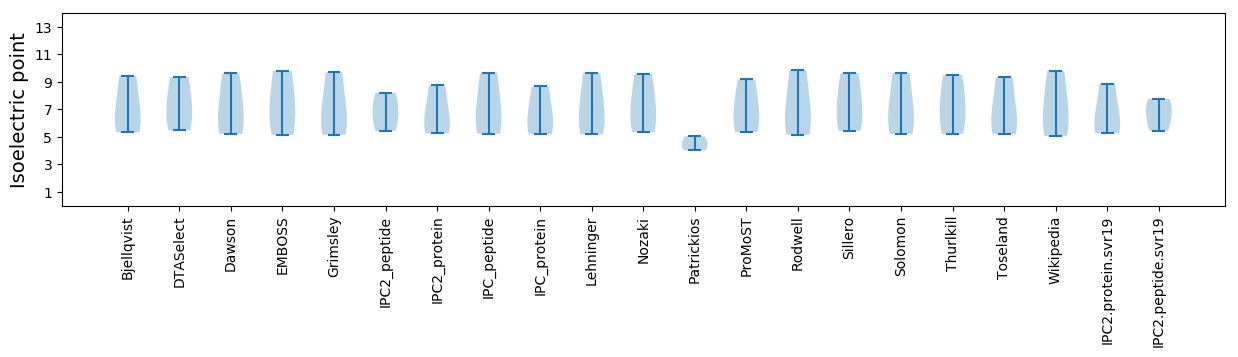
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KG15|A0A1L3KG15_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 22 OX=1923269 PE=4 SV=1
MM1 pKa = 7.78EE2 pKa = 4.72GRR4 pKa = 11.84KK5 pKa = 8.87PRR7 pKa = 11.84AGVRR11 pKa = 11.84RR12 pKa = 11.84EE13 pKa = 3.72SDD15 pKa = 3.14GGSWRR20 pKa = 11.84AHH22 pKa = 3.97VHH24 pKa = 5.35SGEE27 pKa = 3.95ASRR30 pKa = 11.84EE31 pKa = 3.97RR32 pKa = 11.84IEE34 pKa = 5.2DD35 pKa = 3.57VEE37 pKa = 5.14DD38 pKa = 4.41EE39 pKa = 4.09IARR42 pKa = 11.84CEE44 pKa = 4.03EE45 pKa = 3.94ILRR48 pKa = 11.84SSRR51 pKa = 11.84SSWRR55 pKa = 11.84QAAPRR60 pKa = 11.84GAADD64 pKa = 3.74SKK66 pKa = 11.15RR67 pKa = 11.84SGQEE71 pKa = 3.33GRR73 pKa = 11.84RR74 pKa = 11.84SGRR77 pKa = 11.84HH78 pKa = 5.04DD79 pKa = 3.18QGASHH84 pKa = 6.6QGPQGSSSPKK94 pKa = 7.85QAARR98 pKa = 11.84KK99 pKa = 9.47GRR101 pKa = 11.84EE102 pKa = 3.93QEE104 pKa = 5.48DD105 pKa = 3.77PVQPQSSGSSSDD117 pKa = 3.66GEE119 pKa = 4.05EE120 pKa = 4.27GQPPEE125 pKa = 4.55VPALAIPTRR134 pKa = 11.84DD135 pKa = 3.51EE136 pKa = 3.94EE137 pKa = 4.54RR138 pKa = 11.84SEE140 pKa = 4.26SGSGSSGEE148 pKa = 4.17EE149 pKa = 4.13TEE151 pKa = 4.75SEE153 pKa = 4.19GDD155 pKa = 3.73EE156 pKa = 4.4EE157 pKa = 4.21PAQAGQAVQEE167 pKa = 4.64DD168 pKa = 3.85ATEE171 pKa = 4.3PVDD174 pKa = 6.12DD175 pKa = 6.34PDD177 pKa = 4.76DD178 pKa = 3.98CWDD181 pKa = 3.52VGGGANAGCINTGLVNFLKK200 pKa = 10.81AKK202 pKa = 10.07LAYY205 pKa = 9.41KK206 pKa = 10.24PRR208 pKa = 11.84FKK210 pKa = 11.15NGVLDD215 pKa = 3.78QALVNYY221 pKa = 9.83AVAQAQIYY229 pKa = 8.76MRR231 pKa = 11.84DD232 pKa = 3.27HH233 pKa = 7.31PARR236 pKa = 11.84VPEE239 pKa = 4.23DD240 pKa = 3.39KK241 pKa = 11.4YY242 pKa = 11.44EE243 pKa = 4.08VFASSIMQAIRR254 pKa = 11.84DD255 pKa = 3.54ISVEE259 pKa = 4.03EE260 pKa = 3.83NNLRR264 pKa = 11.84KK265 pKa = 10.05LEE267 pKa = 4.2LEE269 pKa = 3.99AAALTRR275 pKa = 11.84HH276 pKa = 5.54TNRR279 pKa = 11.84LTVRR283 pKa = 11.84SKK285 pKa = 11.06EE286 pKa = 3.95FVYY289 pKa = 9.51DD290 pKa = 3.78TGAGRR295 pKa = 11.84FSWRR299 pKa = 11.84YY300 pKa = 6.47WFPKK304 pKa = 9.48ASLVRR309 pKa = 11.84GVPTHH314 pKa = 6.79PIEE317 pKa = 4.32EE318 pKa = 4.5EE319 pKa = 4.06EE320 pKa = 4.29IEE322 pKa = 4.18MGGRR326 pKa = 11.84WVDD329 pKa = 2.99IASVFRR335 pKa = 11.84RR336 pKa = 11.84AIRR339 pKa = 11.84QPIVHH344 pKa = 5.23TTIPRR349 pKa = 11.84LLGQEE354 pKa = 3.93MLGFLAA360 pKa = 5.5
MM1 pKa = 7.78EE2 pKa = 4.72GRR4 pKa = 11.84KK5 pKa = 8.87PRR7 pKa = 11.84AGVRR11 pKa = 11.84RR12 pKa = 11.84EE13 pKa = 3.72SDD15 pKa = 3.14GGSWRR20 pKa = 11.84AHH22 pKa = 3.97VHH24 pKa = 5.35SGEE27 pKa = 3.95ASRR30 pKa = 11.84EE31 pKa = 3.97RR32 pKa = 11.84IEE34 pKa = 5.2DD35 pKa = 3.57VEE37 pKa = 5.14DD38 pKa = 4.41EE39 pKa = 4.09IARR42 pKa = 11.84CEE44 pKa = 4.03EE45 pKa = 3.94ILRR48 pKa = 11.84SSRR51 pKa = 11.84SSWRR55 pKa = 11.84QAAPRR60 pKa = 11.84GAADD64 pKa = 3.74SKK66 pKa = 11.15RR67 pKa = 11.84SGQEE71 pKa = 3.33GRR73 pKa = 11.84RR74 pKa = 11.84SGRR77 pKa = 11.84HH78 pKa = 5.04DD79 pKa = 3.18QGASHH84 pKa = 6.6QGPQGSSSPKK94 pKa = 7.85QAARR98 pKa = 11.84KK99 pKa = 9.47GRR101 pKa = 11.84EE102 pKa = 3.93QEE104 pKa = 5.48DD105 pKa = 3.77PVQPQSSGSSSDD117 pKa = 3.66GEE119 pKa = 4.05EE120 pKa = 4.27GQPPEE125 pKa = 4.55VPALAIPTRR134 pKa = 11.84DD135 pKa = 3.51EE136 pKa = 3.94EE137 pKa = 4.54RR138 pKa = 11.84SEE140 pKa = 4.26SGSGSSGEE148 pKa = 4.17EE149 pKa = 4.13TEE151 pKa = 4.75SEE153 pKa = 4.19GDD155 pKa = 3.73EE156 pKa = 4.4EE157 pKa = 4.21PAQAGQAVQEE167 pKa = 4.64DD168 pKa = 3.85ATEE171 pKa = 4.3PVDD174 pKa = 6.12DD175 pKa = 6.34PDD177 pKa = 4.76DD178 pKa = 3.98CWDD181 pKa = 3.52VGGGANAGCINTGLVNFLKK200 pKa = 10.81AKK202 pKa = 10.07LAYY205 pKa = 9.41KK206 pKa = 10.24PRR208 pKa = 11.84FKK210 pKa = 11.15NGVLDD215 pKa = 3.78QALVNYY221 pKa = 9.83AVAQAQIYY229 pKa = 8.76MRR231 pKa = 11.84DD232 pKa = 3.27HH233 pKa = 7.31PARR236 pKa = 11.84VPEE239 pKa = 4.23DD240 pKa = 3.39KK241 pKa = 11.4YY242 pKa = 11.44EE243 pKa = 4.08VFASSIMQAIRR254 pKa = 11.84DD255 pKa = 3.54ISVEE259 pKa = 4.03EE260 pKa = 3.83NNLRR264 pKa = 11.84KK265 pKa = 10.05LEE267 pKa = 4.2LEE269 pKa = 3.99AAALTRR275 pKa = 11.84HH276 pKa = 5.54TNRR279 pKa = 11.84LTVRR283 pKa = 11.84SKK285 pKa = 11.06EE286 pKa = 3.95FVYY289 pKa = 9.51DD290 pKa = 3.78TGAGRR295 pKa = 11.84FSWRR299 pKa = 11.84YY300 pKa = 6.47WFPKK304 pKa = 9.48ASLVRR309 pKa = 11.84GVPTHH314 pKa = 6.79PIEE317 pKa = 4.32EE318 pKa = 4.5EE319 pKa = 4.06EE320 pKa = 4.29IEE322 pKa = 4.18MGGRR326 pKa = 11.84WVDD329 pKa = 2.99IASVFRR335 pKa = 11.84RR336 pKa = 11.84AIRR339 pKa = 11.84QPIVHH344 pKa = 5.23TTIPRR349 pKa = 11.84LLGQEE354 pKa = 3.93MLGFLAA360 pKa = 5.5
Molecular weight: 39.64 kDa
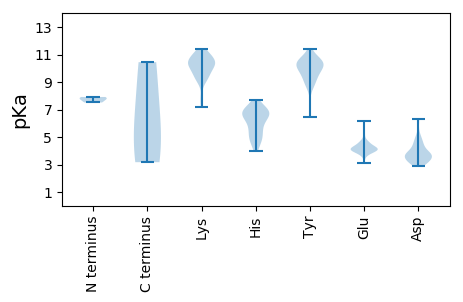
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG15|A0A1L3KG15_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 22 OX=1923269 PE=4 SV=1
MM1 pKa = 7.51FDD3 pKa = 3.05VSRR6 pKa = 11.84ANFSCKK12 pKa = 9.1HH13 pKa = 4.54QRR15 pKa = 11.84EE16 pKa = 4.05MHH18 pKa = 5.73LVEE21 pKa = 4.56DD22 pKa = 3.57HH23 pKa = 6.53VLRR26 pKa = 11.84QEE28 pKa = 3.85RR29 pKa = 11.84FAFHH33 pKa = 6.51NCAPNEE39 pKa = 4.04LDD41 pKa = 3.19SLMRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 6.79VIPQVSPSQEE57 pKa = 3.92YY58 pKa = 10.24IRR60 pKa = 11.84FADD63 pKa = 3.5KK64 pKa = 10.46EE65 pKa = 4.09LRR67 pKa = 11.84ALWRR71 pKa = 11.84SIGAPRR77 pKa = 11.84LSPLDD82 pKa = 4.3DD83 pKa = 4.1EE84 pKa = 5.49SLLSTRR90 pKa = 11.84SRR92 pKa = 11.84HH93 pKa = 4.54VQRR96 pKa = 11.84RR97 pKa = 11.84WKK99 pKa = 10.45RR100 pKa = 11.84IAEE103 pKa = 4.14EE104 pKa = 3.68AATRR108 pKa = 11.84PFSPKK113 pKa = 9.47EE114 pKa = 3.75ALVKK118 pKa = 10.93AFIKK122 pKa = 9.92YY123 pKa = 8.69EE124 pKa = 4.14KK125 pKa = 10.53YY126 pKa = 10.63GLEE129 pKa = 4.0HH130 pKa = 7.71LDD132 pKa = 3.17INKK135 pKa = 9.24PPRR138 pKa = 11.84AIQARR143 pKa = 11.84GPLFTFRR150 pKa = 11.84LAKK153 pKa = 9.6YY154 pKa = 8.92LVPIEE159 pKa = 4.05HH160 pKa = 7.03KK161 pKa = 8.97MWNWKK166 pKa = 9.36PKK168 pKa = 7.74WNHH171 pKa = 4.77GLRR174 pKa = 11.84VFAKK178 pKa = 9.83GRR180 pKa = 11.84NAAEE184 pKa = 4.14RR185 pKa = 11.84AHH187 pKa = 7.13DD188 pKa = 4.44LLKK191 pKa = 10.84LSQVYY196 pKa = 10.34GSATAMIGIDD206 pKa = 3.26HH207 pKa = 6.97SKK209 pKa = 9.92FDD211 pKa = 3.67SRR213 pKa = 11.84VHH215 pKa = 6.43VEE217 pKa = 3.79HH218 pKa = 6.87LKK220 pKa = 10.3ISHH223 pKa = 6.8RR224 pKa = 11.84FNARR228 pKa = 11.84FYY230 pKa = 10.27RR231 pKa = 11.84GQDD234 pKa = 3.1GRR236 pKa = 11.84DD237 pKa = 3.22LSRR240 pKa = 11.84LMRR243 pKa = 11.84CQLKK247 pKa = 10.34NKK249 pKa = 9.72CVTEE253 pKa = 3.75NGIFYY258 pKa = 10.09VVEE261 pKa = 4.11GRR263 pKa = 11.84RR264 pKa = 11.84MSGDD268 pKa = 3.03IDD270 pKa = 3.79TAKK273 pKa = 10.74GNCEE277 pKa = 3.52INLTVLRR284 pKa = 11.84YY285 pKa = 9.28ILRR288 pKa = 11.84GLRR291 pKa = 11.84GAIYY295 pKa = 10.56LDD297 pKa = 4.17GDD299 pKa = 4.04DD300 pKa = 5.08SVVACHH306 pKa = 6.97PDD308 pKa = 3.44DD309 pKa = 3.75VDD311 pKa = 3.56EE312 pKa = 4.26VVRR315 pKa = 11.84RR316 pKa = 11.84VKK318 pKa = 10.7SGAGTGMQSTVEE330 pKa = 4.0VFDD333 pKa = 4.38RR334 pKa = 11.84FSKK337 pKa = 11.36VEE339 pKa = 4.03FCQARR344 pKa = 11.84PIYY347 pKa = 10.49TNGKK351 pKa = 7.18WMMMRR356 pKa = 11.84NPVKK360 pKa = 10.4AISNMCLMMKK370 pKa = 10.37LPEE373 pKa = 4.46EE374 pKa = 4.74GVATRR379 pKa = 11.84LATIGEE385 pKa = 4.66GEE387 pKa = 4.11LHH389 pKa = 6.75ASSGCPTIYY398 pKa = 10.41PFAKK402 pKa = 9.87KK403 pKa = 9.94LRR405 pKa = 11.84RR406 pKa = 11.84HH407 pKa = 5.76GNVDD411 pKa = 3.08EE412 pKa = 5.68AYY414 pKa = 10.27FEE416 pKa = 4.31YY417 pKa = 10.35RR418 pKa = 11.84HH419 pKa = 6.1KK420 pKa = 10.96LNRR423 pKa = 11.84DD424 pKa = 4.73LIPKK428 pKa = 9.73EE429 pKa = 3.96PDD431 pKa = 2.93HH432 pKa = 6.33EE433 pKa = 6.15AEE435 pKa = 4.2ASAWDD440 pKa = 3.77AWDD443 pKa = 3.76LPPYY447 pKa = 10.02YY448 pKa = 10.47NRR450 pKa = 11.84LSVV453 pKa = 3.17
MM1 pKa = 7.51FDD3 pKa = 3.05VSRR6 pKa = 11.84ANFSCKK12 pKa = 9.1HH13 pKa = 4.54QRR15 pKa = 11.84EE16 pKa = 4.05MHH18 pKa = 5.73LVEE21 pKa = 4.56DD22 pKa = 3.57HH23 pKa = 6.53VLRR26 pKa = 11.84QEE28 pKa = 3.85RR29 pKa = 11.84FAFHH33 pKa = 6.51NCAPNEE39 pKa = 4.04LDD41 pKa = 3.19SLMRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 6.79VIPQVSPSQEE57 pKa = 3.92YY58 pKa = 10.24IRR60 pKa = 11.84FADD63 pKa = 3.5KK64 pKa = 10.46EE65 pKa = 4.09LRR67 pKa = 11.84ALWRR71 pKa = 11.84SIGAPRR77 pKa = 11.84LSPLDD82 pKa = 4.3DD83 pKa = 4.1EE84 pKa = 5.49SLLSTRR90 pKa = 11.84SRR92 pKa = 11.84HH93 pKa = 4.54VQRR96 pKa = 11.84RR97 pKa = 11.84WKK99 pKa = 10.45RR100 pKa = 11.84IAEE103 pKa = 4.14EE104 pKa = 3.68AATRR108 pKa = 11.84PFSPKK113 pKa = 9.47EE114 pKa = 3.75ALVKK118 pKa = 10.93AFIKK122 pKa = 9.92YY123 pKa = 8.69EE124 pKa = 4.14KK125 pKa = 10.53YY126 pKa = 10.63GLEE129 pKa = 4.0HH130 pKa = 7.71LDD132 pKa = 3.17INKK135 pKa = 9.24PPRR138 pKa = 11.84AIQARR143 pKa = 11.84GPLFTFRR150 pKa = 11.84LAKK153 pKa = 9.6YY154 pKa = 8.92LVPIEE159 pKa = 4.05HH160 pKa = 7.03KK161 pKa = 8.97MWNWKK166 pKa = 9.36PKK168 pKa = 7.74WNHH171 pKa = 4.77GLRR174 pKa = 11.84VFAKK178 pKa = 9.83GRR180 pKa = 11.84NAAEE184 pKa = 4.14RR185 pKa = 11.84AHH187 pKa = 7.13DD188 pKa = 4.44LLKK191 pKa = 10.84LSQVYY196 pKa = 10.34GSATAMIGIDD206 pKa = 3.26HH207 pKa = 6.97SKK209 pKa = 9.92FDD211 pKa = 3.67SRR213 pKa = 11.84VHH215 pKa = 6.43VEE217 pKa = 3.79HH218 pKa = 6.87LKK220 pKa = 10.3ISHH223 pKa = 6.8RR224 pKa = 11.84FNARR228 pKa = 11.84FYY230 pKa = 10.27RR231 pKa = 11.84GQDD234 pKa = 3.1GRR236 pKa = 11.84DD237 pKa = 3.22LSRR240 pKa = 11.84LMRR243 pKa = 11.84CQLKK247 pKa = 10.34NKK249 pKa = 9.72CVTEE253 pKa = 3.75NGIFYY258 pKa = 10.09VVEE261 pKa = 4.11GRR263 pKa = 11.84RR264 pKa = 11.84MSGDD268 pKa = 3.03IDD270 pKa = 3.79TAKK273 pKa = 10.74GNCEE277 pKa = 3.52INLTVLRR284 pKa = 11.84YY285 pKa = 9.28ILRR288 pKa = 11.84GLRR291 pKa = 11.84GAIYY295 pKa = 10.56LDD297 pKa = 4.17GDD299 pKa = 4.04DD300 pKa = 5.08SVVACHH306 pKa = 6.97PDD308 pKa = 3.44DD309 pKa = 3.75VDD311 pKa = 3.56EE312 pKa = 4.26VVRR315 pKa = 11.84RR316 pKa = 11.84VKK318 pKa = 10.7SGAGTGMQSTVEE330 pKa = 4.0VFDD333 pKa = 4.38RR334 pKa = 11.84FSKK337 pKa = 11.36VEE339 pKa = 4.03FCQARR344 pKa = 11.84PIYY347 pKa = 10.49TNGKK351 pKa = 7.18WMMMRR356 pKa = 11.84NPVKK360 pKa = 10.4AISNMCLMMKK370 pKa = 10.37LPEE373 pKa = 4.46EE374 pKa = 4.74GVATRR379 pKa = 11.84LATIGEE385 pKa = 4.66GEE387 pKa = 4.11LHH389 pKa = 6.75ASSGCPTIYY398 pKa = 10.41PFAKK402 pKa = 9.87KK403 pKa = 9.94LRR405 pKa = 11.84RR406 pKa = 11.84HH407 pKa = 5.76GNVDD411 pKa = 3.08EE412 pKa = 5.68AYY414 pKa = 10.27FEE416 pKa = 4.31YY417 pKa = 10.35RR418 pKa = 11.84HH419 pKa = 6.1KK420 pKa = 10.96LNRR423 pKa = 11.84DD424 pKa = 4.73LIPKK428 pKa = 9.73EE429 pKa = 3.96PDD431 pKa = 2.93HH432 pKa = 6.33EE433 pKa = 6.15AEE435 pKa = 4.2ASAWDD440 pKa = 3.77AWDD443 pKa = 3.76LPPYY447 pKa = 10.02YY448 pKa = 10.47NRR450 pKa = 11.84LSVV453 pKa = 3.17
Molecular weight: 52.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213 |
360 |
453 |
404.3 |
45.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.574 ± 0.584 | 1.566 ± 0.306 |
5.276 ± 0.429 | 7.749 ± 1.265 |
3.38 ± 0.505 | 7.749 ± 1.058 |
2.721 ± 0.693 | 4.452 ± 0.122 |
4.782 ± 0.725 | 6.843 ± 0.935 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.431 | 3.545 ± 0.545 |
5.688 ± 0.408 | 3.133 ± 0.883 |
8.574 ± 1.176 | 7.42 ± 0.812 |
4.617 ± 1.406 | 6.678 ± 0.354 |
1.731 ± 0.027 | 3.215 ± 0.695 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
