
Heliocybe sulcata
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Gloeophyllales; Gloeophyllales incertae sedis; Heliocybe
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
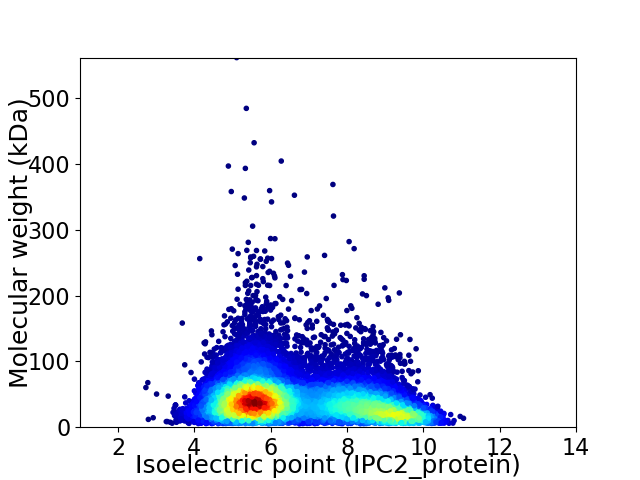
Virtual 2D-PAGE plot for 12554 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C3NDQ9|A0A5C3NDQ9_9AGAM Glycoside hydrolase family 105 protein OS=Heliocybe sulcata OX=5364 GN=OE88DRAFT_1672797 PE=4 SV=1
DDD2 pKa = 3.37SYYY5 pKa = 10.51GSISIGSPAQQFNVILDDD23 pKa = 3.74GSADDD28 pKa = 3.07WVAGSSCSSCSSSTPTFSTSRR49 pKa = 11.84SSSFQSSTSSAGQQQSLTIRR69 pKa = 11.84YYY71 pKa = 9.1SGQVTGSLGADDD83 pKa = 3.55VQMAGFQVSGQEEE96 pKa = 4.05VVVDDD101 pKa = 3.68TSQNLLSGSNAGIMGLGFQAIASSGATPFWEEE133 pKa = 4.1LVNANQLSQPLMAFWLRR150 pKa = 11.84RR151 pKa = 11.84LLDDD155 pKa = 4.44DD156 pKa = 5.32SATQNSEEE164 pKa = 4.02YYY166 pKa = 10.65GEEE169 pKa = 3.94TLGGTNTSLYYY180 pKa = 9.95GDDD183 pKa = 3.21QFIDDD188 pKa = 4.38PSGVQQSFWLLEEE201 pKa = 4.02QGVSVQGGSVGISTGNAALAAIDDD225 pKa = 3.72GTTLVGGPSNDDD237 pKa = 3.31DDD239 pKa = 4.93IWAAVSGSQALSGQYYY255 pKa = 10.56GFYYY259 pKa = 10.43FPCSTNLQVSLSFGGNAWPISPADDD284 pKa = 3.8NLGAISGNMCLGGIFDDD301 pKa = 4.72TQGADDD307 pKa = 3.27GSGNPGWVVGDDD319 pKa = 3.66FLKKK323 pKa = 10.46VYYY326 pKa = 8.57VFRR329 pKa = 11.84ASPASVGFAQLSEEE343 pKa = 4.13AGGSSGSSPHHH354 pKa = 6.08AASSSSARR362 pKa = 11.84S
DDD2 pKa = 3.37SYYY5 pKa = 10.51GSISIGSPAQQFNVILDDD23 pKa = 3.74GSADDD28 pKa = 3.07WVAGSSCSSCSSSTPTFSTSRR49 pKa = 11.84SSSFQSSTSSAGQQQSLTIRR69 pKa = 11.84YYY71 pKa = 9.1SGQVTGSLGADDD83 pKa = 3.55VQMAGFQVSGQEEE96 pKa = 4.05VVVDDD101 pKa = 3.68TSQNLLSGSNAGIMGLGFQAIASSGATPFWEEE133 pKa = 4.1LVNANQLSQPLMAFWLRR150 pKa = 11.84RR151 pKa = 11.84LLDDD155 pKa = 4.44DD156 pKa = 5.32SATQNSEEE164 pKa = 4.02YYY166 pKa = 10.65GEEE169 pKa = 3.94TLGGTNTSLYYY180 pKa = 9.95GDDD183 pKa = 3.21QFIDDD188 pKa = 4.38PSGVQQSFWLLEEE201 pKa = 4.02QGVSVQGGSVGISTGNAALAAIDDD225 pKa = 3.72GTTLVGGPSNDDD237 pKa = 3.31DDD239 pKa = 4.93IWAAVSGSQALSGQYYY255 pKa = 10.56GFYYY259 pKa = 10.43FPCSTNLQVSLSFGGNAWPISPADDD284 pKa = 3.8NLGAISGNMCLGGIFDDD301 pKa = 4.72TQGADDD307 pKa = 3.27GSGNPGWVVGDDD319 pKa = 3.66FLKKK323 pKa = 10.46VYYY326 pKa = 8.57VFRR329 pKa = 11.84ASPASVGFAQLSEEE343 pKa = 4.13AGGSSGSSPHHH354 pKa = 6.08AASSSSARR362 pKa = 11.84S
Molecular weight: 36.84 kDa
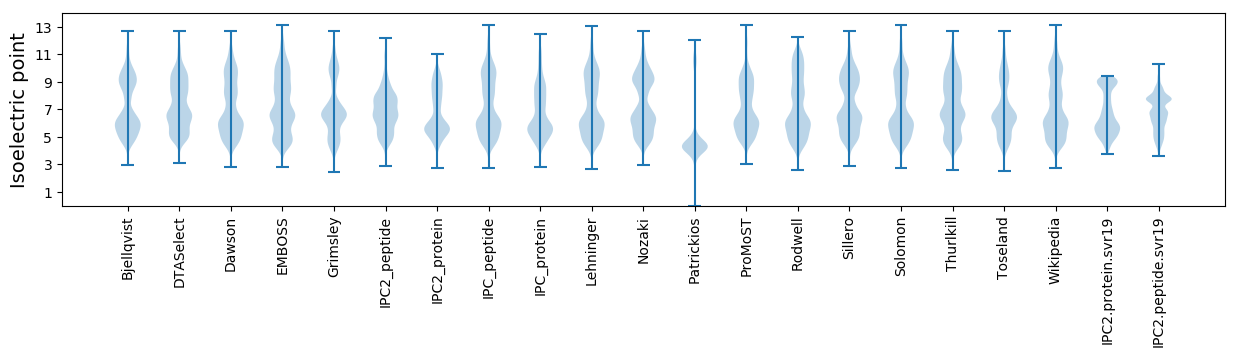
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C3NMN1|A0A5C3NMN1_9AGAM Uncharacterized protein OS=Heliocybe sulcata OX=5364 GN=OE88DRAFT_973144 PE=4 SV=1
MM1 pKa = 7.55PRR3 pKa = 11.84ILNQIVQLLARR14 pKa = 11.84PPTLPRR20 pKa = 11.84PSSAVSALAQLHH32 pKa = 5.34RR33 pKa = 11.84TSLQRR38 pKa = 11.84TTLLPFTSPFQSSSALLAPSFVGQPSALLQLTQVRR73 pKa = 11.84WAARR77 pKa = 11.84GTEE80 pKa = 4.11YY81 pKa = 10.77QPSQRR86 pKa = 11.84VRR88 pKa = 11.84KK89 pKa = 9.28RR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 4.84GFLARR97 pKa = 11.84KK98 pKa = 9.55RR99 pKa = 11.84SLHH102 pKa = 4.21GHH104 pKa = 6.86KK105 pKa = 10.1ILARR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84AKK113 pKa = 9.85GRR115 pKa = 11.84KK116 pKa = 8.43FLSHH120 pKa = 6.98
MM1 pKa = 7.55PRR3 pKa = 11.84ILNQIVQLLARR14 pKa = 11.84PPTLPRR20 pKa = 11.84PSSAVSALAQLHH32 pKa = 5.34RR33 pKa = 11.84TSLQRR38 pKa = 11.84TTLLPFTSPFQSSSALLAPSFVGQPSALLQLTQVRR73 pKa = 11.84WAARR77 pKa = 11.84GTEE80 pKa = 4.11YY81 pKa = 10.77QPSQRR86 pKa = 11.84VRR88 pKa = 11.84KK89 pKa = 9.28RR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 4.84GFLARR97 pKa = 11.84KK98 pKa = 9.55RR99 pKa = 11.84SLHH102 pKa = 4.21GHH104 pKa = 6.86KK105 pKa = 10.1ILARR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84AKK113 pKa = 9.85GRR115 pKa = 11.84KK116 pKa = 8.43FLSHH120 pKa = 6.98
Molecular weight: 13.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5216656 |
49 |
5043 |
415.5 |
45.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.806 ± 0.02 | 1.315 ± 0.009 |
5.537 ± 0.016 | 6.043 ± 0.023 |
3.531 ± 0.014 | 6.727 ± 0.022 |
2.488 ± 0.011 | 4.576 ± 0.014 |
4.418 ± 0.023 | 9.22 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.009 | 3.144 ± 0.011 |
6.522 ± 0.027 | 3.673 ± 0.013 |
6.484 ± 0.02 | 8.694 ± 0.031 |
5.841 ± 0.015 | 6.507 ± 0.018 |
1.507 ± 0.008 | 2.784 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
