
Mesorhizobium amorphae CCNWGS0123
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; Mesorhizobium amorphae
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
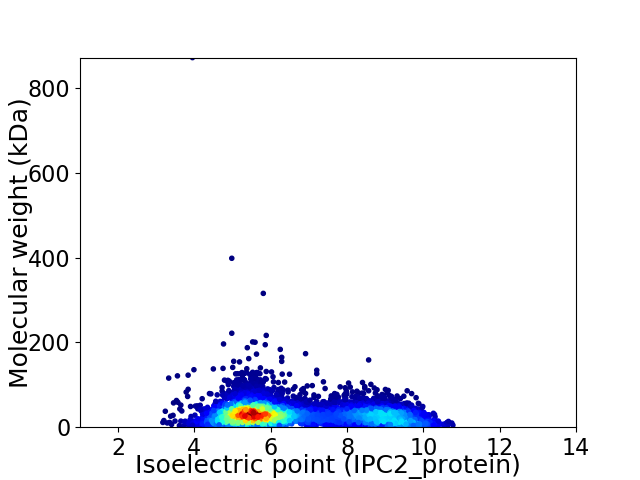
Virtual 2D-PAGE plot for 7083 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
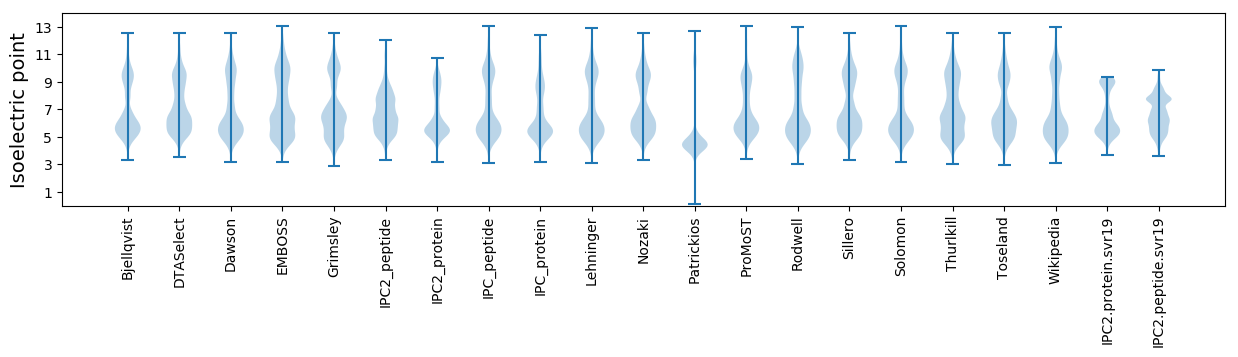
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G6YGA2|G6YGA2_9RHIZ Transposase IS116/IS110/IS902 OS=Mesorhizobium amorphae CCNWGS0123 OX=1082933 GN=MEA186_25052 PE=4 SV=1
MM1 pKa = 7.25ATQIAGAGGTTTSFSNTPQAKK22 pKa = 10.03DD23 pKa = 3.35DD24 pKa = 3.8VFNYY28 pKa = 10.35TEE30 pKa = 4.05DD31 pKa = 3.43TVVIVSAAQSIILLDD46 pKa = 3.59VMKK49 pKa = 10.98NDD51 pKa = 4.04LGGNAKK57 pKa = 8.33TLYY60 pKa = 10.62SVDD63 pKa = 4.16DD64 pKa = 5.12GISASTATKK73 pKa = 10.0QYY75 pKa = 11.53APIDD79 pKa = 4.01LTTADD84 pKa = 3.78AQVSGVSAWEE94 pKa = 4.58DD95 pKa = 2.84IGGGVFIRR103 pKa = 11.84INNGKK108 pKa = 9.47VEE110 pKa = 4.34MNLSQYY116 pKa = 11.04LVLHH120 pKa = 6.51GFSSLQALGAGDD132 pKa = 4.74SINEE136 pKa = 4.16TFTYY140 pKa = 10.09AIKK143 pKa = 10.66LGNGTLSWASVSVNIQGSNDD163 pKa = 3.2GATITAAPGADD174 pKa = 3.12KK175 pKa = 10.67TVVEE179 pKa = 4.76AGGVANGTLNDD190 pKa = 4.34PSAQGQLILTDD201 pKa = 3.47IDD203 pKa = 3.55AGQNHH208 pKa = 5.94FQDD211 pKa = 4.19PASLQGSYY219 pKa = 9.34GTFTFDD225 pKa = 2.91TTTGAWTYY233 pKa = 11.72ALNQALADD241 pKa = 4.32PLTQGQAVTDD251 pKa = 3.99TLTVTSADD259 pKa = 3.08GTASYY264 pKa = 10.76NIVVNITGTNDD275 pKa = 3.33AAVLSADD282 pKa = 3.34VRR284 pKa = 11.84NLTEE288 pKa = 4.81GDD290 pKa = 3.35TAADD294 pKa = 3.28ISTSGTLTISDD305 pKa = 3.58VDD307 pKa = 3.9SPEE310 pKa = 3.94TFVAQVGTVGSYY322 pKa = 8.92GTFDD326 pKa = 3.72IDD328 pKa = 4.75SAGAWTYY335 pKa = 8.06TASSAHH341 pKa = 5.94NEE343 pKa = 4.1FVAGQHH349 pKa = 4.94YY350 pKa = 10.96VEE352 pKa = 5.3DD353 pKa = 4.65FDD355 pKa = 5.33VVSADD360 pKa = 3.51GTHH363 pKa = 5.25TTVHH367 pKa = 6.59IDD369 pKa = 2.98ILGG372 pKa = 3.59
MM1 pKa = 7.25ATQIAGAGGTTTSFSNTPQAKK22 pKa = 10.03DD23 pKa = 3.35DD24 pKa = 3.8VFNYY28 pKa = 10.35TEE30 pKa = 4.05DD31 pKa = 3.43TVVIVSAAQSIILLDD46 pKa = 3.59VMKK49 pKa = 10.98NDD51 pKa = 4.04LGGNAKK57 pKa = 8.33TLYY60 pKa = 10.62SVDD63 pKa = 4.16DD64 pKa = 5.12GISASTATKK73 pKa = 10.0QYY75 pKa = 11.53APIDD79 pKa = 4.01LTTADD84 pKa = 3.78AQVSGVSAWEE94 pKa = 4.58DD95 pKa = 2.84IGGGVFIRR103 pKa = 11.84INNGKK108 pKa = 9.47VEE110 pKa = 4.34MNLSQYY116 pKa = 11.04LVLHH120 pKa = 6.51GFSSLQALGAGDD132 pKa = 4.74SINEE136 pKa = 4.16TFTYY140 pKa = 10.09AIKK143 pKa = 10.66LGNGTLSWASVSVNIQGSNDD163 pKa = 3.2GATITAAPGADD174 pKa = 3.12KK175 pKa = 10.67TVVEE179 pKa = 4.76AGGVANGTLNDD190 pKa = 4.34PSAQGQLILTDD201 pKa = 3.47IDD203 pKa = 3.55AGQNHH208 pKa = 5.94FQDD211 pKa = 4.19PASLQGSYY219 pKa = 9.34GTFTFDD225 pKa = 2.91TTTGAWTYY233 pKa = 11.72ALNQALADD241 pKa = 4.32PLTQGQAVTDD251 pKa = 3.99TLTVTSADD259 pKa = 3.08GTASYY264 pKa = 10.76NIVVNITGTNDD275 pKa = 3.33AAVLSADD282 pKa = 3.34VRR284 pKa = 11.84NLTEE288 pKa = 4.81GDD290 pKa = 3.35TAADD294 pKa = 3.28ISTSGTLTISDD305 pKa = 3.58VDD307 pKa = 3.9SPEE310 pKa = 3.94TFVAQVGTVGSYY322 pKa = 8.92GTFDD326 pKa = 3.72IDD328 pKa = 4.75SAGAWTYY335 pKa = 8.06TASSAHH341 pKa = 5.94NEE343 pKa = 4.1FVAGQHH349 pKa = 4.94YY350 pKa = 10.96VEE352 pKa = 5.3DD353 pKa = 4.65FDD355 pKa = 5.33VVSADD360 pKa = 3.51GTHH363 pKa = 5.25TTVHH367 pKa = 6.59IDD369 pKa = 2.98ILGG372 pKa = 3.59
Molecular weight: 38.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G6Y9T3|G6Y9T3_9RHIZ DNA-directed RNA polymerase subunit omega OS=Mesorhizobium amorphae CCNWGS0123 OX=1082933 GN=rpoZ PE=3 SV=1
MM1 pKa = 7.67SKK3 pKa = 8.45ITLRR7 pKa = 11.84AAVCRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GFFLGNAFIFPGAAPANLRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84IIHH39 pKa = 5.57NRR41 pKa = 11.84RR42 pKa = 3.12
MM1 pKa = 7.67SKK3 pKa = 8.45ITLRR7 pKa = 11.84AAVCRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GFFLGNAFIFPGAAPANLRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84IIHH39 pKa = 5.57NRR41 pKa = 11.84RR42 pKa = 3.12
Molecular weight: 4.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2092589 |
25 |
8568 |
295.4 |
32.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.403 ± 0.037 | 0.832 ± 0.009 |
5.739 ± 0.025 | 5.559 ± 0.026 |
3.892 ± 0.02 | 8.552 ± 0.038 |
2.046 ± 0.014 | 5.429 ± 0.023 |
3.68 ± 0.025 | 9.911 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.508 ± 0.017 | 2.701 ± 0.019 |
5.007 ± 0.023 | 3.042 ± 0.018 |
6.821 ± 0.032 | 5.581 ± 0.025 |
5.252 ± 0.026 | 7.474 ± 0.022 |
1.341 ± 0.014 | 2.231 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
