
Gluconobacter morbifer G707
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Gluconobacter; Gluconobacter morbifer
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
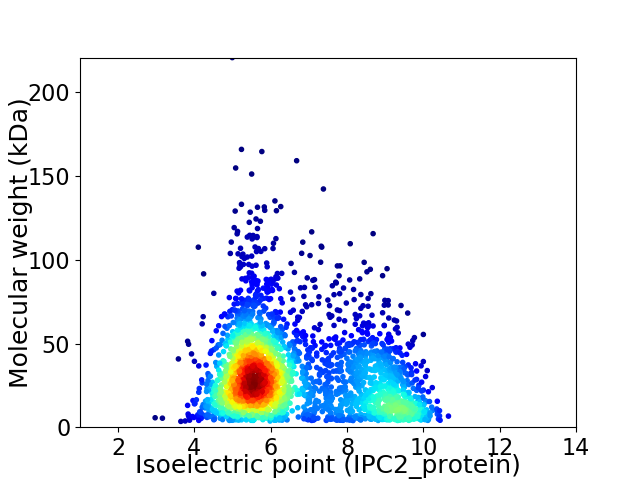
Virtual 2D-PAGE plot for 2846 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G6XKP4|G6XKP4_9PROT CMD domain-containing protein OS=Gluconobacter morbifer G707 OX=1088869 GN=GMO_20600 PE=4 SV=1
MM1 pKa = 7.69AGLKK5 pKa = 10.39SSAHH9 pKa = 6.12YY10 pKa = 10.92DD11 pKa = 3.39LTSGSNSITGGSAAQGLISSGGYY34 pKa = 6.83YY35 pKa = 9.82TINGEE40 pKa = 4.32LGYY43 pKa = 10.03IAAGSYY49 pKa = 10.89GDD51 pKa = 4.0SSNPQDD57 pKa = 3.61TLNSGVAIDD66 pKa = 4.73LRR68 pKa = 11.84NNTASSVSVLAGDD81 pKa = 3.61QAGVTVYY88 pKa = 10.82AGDD91 pKa = 3.46QSGSFVGGLGDD102 pKa = 4.16NVFLGSGKK110 pKa = 8.31TGSWNVATGSGNDD123 pKa = 4.06TILGTNGNSTIDD135 pKa = 3.64GGTGDD140 pKa = 3.58NLIYY144 pKa = 10.53LGSGTNVVRR153 pKa = 11.84SEE155 pKa = 4.15GQDD158 pKa = 3.39TIDD161 pKa = 3.8GGGGVDD167 pKa = 3.4TVTLLGGSSVVSLQNNATVYY187 pKa = 8.83DD188 pKa = 4.14TTGHH192 pKa = 6.33NDD194 pKa = 2.99VTVGSNSSITGGSSSTYY211 pKa = 8.28FTTGSMSTISGGQNDD226 pKa = 4.98TISASGDD233 pKa = 3.29LEE235 pKa = 4.17QIRR238 pKa = 11.84GSGNNLSVGGSLTFLNGTGSTTITAGNATLFGASGQDD275 pKa = 2.97IQYY278 pKa = 9.35TGTSGTALYY287 pKa = 10.55VAGDD291 pKa = 3.68GSEE294 pKa = 4.49TIDD297 pKa = 3.49ASASKK302 pKa = 9.03TAINAFAGTGDD313 pKa = 3.58DD314 pKa = 4.13TIIGGSAADD323 pKa = 3.71TMVGGSGNATLTGGSGAANLFALVDD348 pKa = 4.32GKK350 pKa = 11.01AGGDD354 pKa = 3.47YY355 pKa = 10.49TITDD359 pKa = 4.54FGSAAGNLVALYY371 pKa = 10.67NYY373 pKa = 9.63GLNSNTLQTVLNDD386 pKa = 3.41ATVSGGNTTIALSDD400 pKa = 3.36NSKK403 pKa = 8.49ITFVGVTDD411 pKa = 4.97LKK413 pKa = 10.43TSNFTGG419 pKa = 3.49
MM1 pKa = 7.69AGLKK5 pKa = 10.39SSAHH9 pKa = 6.12YY10 pKa = 10.92DD11 pKa = 3.39LTSGSNSITGGSAAQGLISSGGYY34 pKa = 6.83YY35 pKa = 9.82TINGEE40 pKa = 4.32LGYY43 pKa = 10.03IAAGSYY49 pKa = 10.89GDD51 pKa = 4.0SSNPQDD57 pKa = 3.61TLNSGVAIDD66 pKa = 4.73LRR68 pKa = 11.84NNTASSVSVLAGDD81 pKa = 3.61QAGVTVYY88 pKa = 10.82AGDD91 pKa = 3.46QSGSFVGGLGDD102 pKa = 4.16NVFLGSGKK110 pKa = 8.31TGSWNVATGSGNDD123 pKa = 4.06TILGTNGNSTIDD135 pKa = 3.64GGTGDD140 pKa = 3.58NLIYY144 pKa = 10.53LGSGTNVVRR153 pKa = 11.84SEE155 pKa = 4.15GQDD158 pKa = 3.39TIDD161 pKa = 3.8GGGGVDD167 pKa = 3.4TVTLLGGSSVVSLQNNATVYY187 pKa = 8.83DD188 pKa = 4.14TTGHH192 pKa = 6.33NDD194 pKa = 2.99VTVGSNSSITGGSSSTYY211 pKa = 8.28FTTGSMSTISGGQNDD226 pKa = 4.98TISASGDD233 pKa = 3.29LEE235 pKa = 4.17QIRR238 pKa = 11.84GSGNNLSVGGSLTFLNGTGSTTITAGNATLFGASGQDD275 pKa = 2.97IQYY278 pKa = 9.35TGTSGTALYY287 pKa = 10.55VAGDD291 pKa = 3.68GSEE294 pKa = 4.49TIDD297 pKa = 3.49ASASKK302 pKa = 9.03TAINAFAGTGDD313 pKa = 3.58DD314 pKa = 4.13TIIGGSAADD323 pKa = 3.71TMVGGSGNATLTGGSGAANLFALVDD348 pKa = 4.32GKK350 pKa = 11.01AGGDD354 pKa = 3.47YY355 pKa = 10.49TITDD359 pKa = 4.54FGSAAGNLVALYY371 pKa = 10.67NYY373 pKa = 9.63GLNSNTLQTVLNDD386 pKa = 3.41ATVSGGNTTIALSDD400 pKa = 3.36NSKK403 pKa = 8.49ITFVGVTDD411 pKa = 4.97LKK413 pKa = 10.43TSNFTGG419 pKa = 3.49
Molecular weight: 40.78 kDa
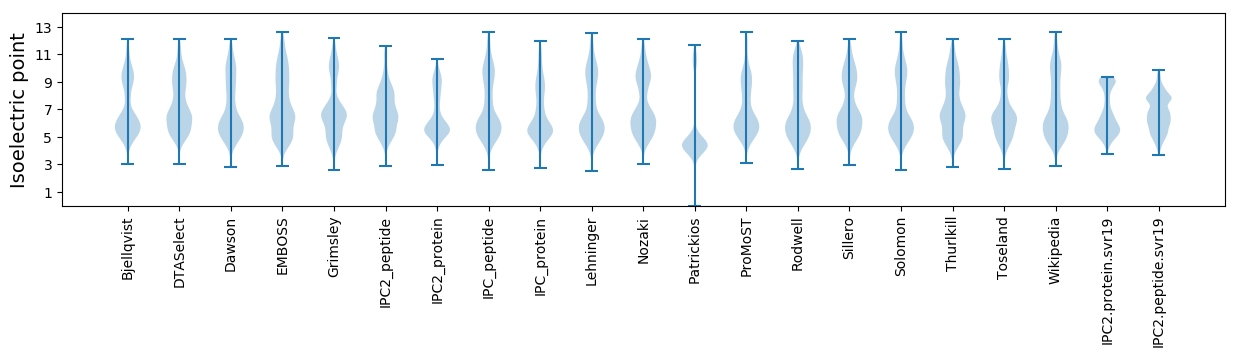
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G6XH41|G6XH41_9PROT Glycerol-3-phosphate dehydrogenase OS=Gluconobacter morbifer G707 OX=1088869 GN=GMO_08060 PE=3 SV=1
MM1 pKa = 7.63PRR3 pKa = 11.84PSGFSMPDD11 pKa = 2.99ALEE14 pKa = 3.6QRR16 pKa = 11.84LIGEE20 pKa = 4.73LTTAVDD26 pKa = 3.6PAVVAFAHH34 pKa = 6.31NLAQSMPAPPLGVLFYY50 pKa = 11.45GSMLRR55 pKa = 11.84SADD58 pKa = 3.41PDD60 pKa = 4.65GILDD64 pKa = 3.9FYY66 pKa = 10.9IITEE70 pKa = 4.15TARR73 pKa = 11.84DD74 pKa = 3.94FPGGFLARR82 pKa = 11.84AANRR86 pKa = 11.84LLPPNVRR93 pKa = 11.84YY94 pKa = 10.34AEE96 pKa = 4.26FTASDD101 pKa = 3.55NRR103 pKa = 11.84ILRR106 pKa = 11.84AKK108 pKa = 10.11IAILSRR114 pKa = 11.84AQFAARR120 pKa = 11.84TSLASLDD127 pKa = 3.53TTLWARR133 pKa = 11.84FCQPARR139 pKa = 11.84LIWVRR144 pKa = 11.84GPEE147 pKa = 3.98AADD150 pKa = 3.48ALLDD154 pKa = 4.53LIARR158 pKa = 11.84CVTTAACWSALLGEE172 pKa = 4.64PRR174 pKa = 11.84MTALGFWRR182 pKa = 11.84TLFAQTYY189 pKa = 8.23ASEE192 pKa = 4.13LRR194 pKa = 11.84VEE196 pKa = 4.32KK197 pKa = 10.51KK198 pKa = 10.83GRR200 pKa = 11.84GNSLLQGQEE209 pKa = 3.29ARR211 pKa = 11.84YY212 pKa = 9.01AALLALGWARR222 pKa = 11.84AHH224 pKa = 6.18LQFSADD230 pKa = 3.61GQLLEE235 pKa = 4.18PVIPRR240 pKa = 11.84EE241 pKa = 4.06LRR243 pKa = 11.84LKK245 pKa = 10.48AVQRR249 pKa = 11.84WQLVRR254 pKa = 11.84RR255 pKa = 11.84TGQPRR260 pKa = 11.84NVMRR264 pKa = 11.84LLKK267 pKa = 10.67GAFTFEE273 pKa = 4.08NGASYY278 pKa = 10.1LAWKK282 pKa = 9.51IRR284 pKa = 11.84RR285 pKa = 11.84HH286 pKa = 5.11TGFDD290 pKa = 3.16LRR292 pKa = 11.84LSAFEE297 pKa = 3.93QRR299 pKa = 11.84HH300 pKa = 5.63PLLMLPALLWRR311 pKa = 11.84ARR313 pKa = 11.84KK314 pKa = 8.72LWNRR318 pKa = 11.84PQDD321 pKa = 3.27GMVPEE326 pKa = 4.97KK327 pKa = 11.14KK328 pKa = 10.33NGTQQ332 pKa = 3.1
MM1 pKa = 7.63PRR3 pKa = 11.84PSGFSMPDD11 pKa = 2.99ALEE14 pKa = 3.6QRR16 pKa = 11.84LIGEE20 pKa = 4.73LTTAVDD26 pKa = 3.6PAVVAFAHH34 pKa = 6.31NLAQSMPAPPLGVLFYY50 pKa = 11.45GSMLRR55 pKa = 11.84SADD58 pKa = 3.41PDD60 pKa = 4.65GILDD64 pKa = 3.9FYY66 pKa = 10.9IITEE70 pKa = 4.15TARR73 pKa = 11.84DD74 pKa = 3.94FPGGFLARR82 pKa = 11.84AANRR86 pKa = 11.84LLPPNVRR93 pKa = 11.84YY94 pKa = 10.34AEE96 pKa = 4.26FTASDD101 pKa = 3.55NRR103 pKa = 11.84ILRR106 pKa = 11.84AKK108 pKa = 10.11IAILSRR114 pKa = 11.84AQFAARR120 pKa = 11.84TSLASLDD127 pKa = 3.53TTLWARR133 pKa = 11.84FCQPARR139 pKa = 11.84LIWVRR144 pKa = 11.84GPEE147 pKa = 3.98AADD150 pKa = 3.48ALLDD154 pKa = 4.53LIARR158 pKa = 11.84CVTTAACWSALLGEE172 pKa = 4.64PRR174 pKa = 11.84MTALGFWRR182 pKa = 11.84TLFAQTYY189 pKa = 8.23ASEE192 pKa = 4.13LRR194 pKa = 11.84VEE196 pKa = 4.32KK197 pKa = 10.51KK198 pKa = 10.83GRR200 pKa = 11.84GNSLLQGQEE209 pKa = 3.29ARR211 pKa = 11.84YY212 pKa = 9.01AALLALGWARR222 pKa = 11.84AHH224 pKa = 6.18LQFSADD230 pKa = 3.61GQLLEE235 pKa = 4.18PVIPRR240 pKa = 11.84EE241 pKa = 4.06LRR243 pKa = 11.84LKK245 pKa = 10.48AVQRR249 pKa = 11.84WQLVRR254 pKa = 11.84RR255 pKa = 11.84TGQPRR260 pKa = 11.84NVMRR264 pKa = 11.84LLKK267 pKa = 10.67GAFTFEE273 pKa = 4.08NGASYY278 pKa = 10.1LAWKK282 pKa = 9.51IRR284 pKa = 11.84RR285 pKa = 11.84HH286 pKa = 5.11TGFDD290 pKa = 3.16LRR292 pKa = 11.84LSAFEE297 pKa = 3.93QRR299 pKa = 11.84HH300 pKa = 5.63PLLMLPALLWRR311 pKa = 11.84ARR313 pKa = 11.84KK314 pKa = 8.72LWNRR318 pKa = 11.84PQDD321 pKa = 3.27GMVPEE326 pKa = 4.97KK327 pKa = 11.14KK328 pKa = 10.33NGTQQ332 pKa = 3.1
Molecular weight: 37.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
857605 |
37 |
1992 |
301.3 |
32.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.011 ± 0.058 | 1.032 ± 0.016 |
5.598 ± 0.034 | 5.324 ± 0.044 |
3.567 ± 0.03 | 8.416 ± 0.044 |
2.467 ± 0.025 | 4.971 ± 0.035 |
2.914 ± 0.033 | 10.464 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.581 ± 0.022 | 2.773 ± 0.034 |
5.583 ± 0.04 | 3.583 ± 0.032 |
7.239 ± 0.054 | 6.017 ± 0.042 |
5.816 ± 0.037 | 7.023 ± 0.042 |
1.385 ± 0.02 | 2.237 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
