
Theileria annulata
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Piroplasmida; Theileriidae; Theileria
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
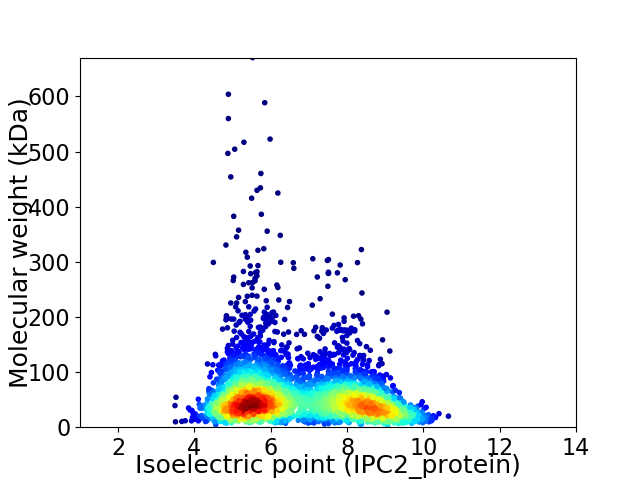
Virtual 2D-PAGE plot for 3790 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4UIL9|Q4UIL9_THEAN Uncharacterized protein OS=Theileria annulata OX=5874 GN=TA16905 PE=4 SV=1
MM1 pKa = 7.29NLIITLFAILIHH13 pKa = 6.65VSICLDD19 pKa = 3.47PVDD22 pKa = 5.53IEE24 pKa = 4.5LLQPLPEE31 pKa = 4.22EE32 pKa = 5.07GIVSEE37 pKa = 4.44EE38 pKa = 4.13VTFHH42 pKa = 6.89GIHH45 pKa = 5.53YY46 pKa = 9.47QIIVPLGQYY55 pKa = 7.65YY56 pKa = 9.95FRR58 pKa = 11.84TISYY62 pKa = 9.66NNNPIFQEE70 pKa = 4.0AQRR73 pKa = 11.84ACLKK77 pKa = 10.56VVIVTSDD84 pKa = 3.3HH85 pKa = 5.96IKK87 pKa = 10.87YY88 pKa = 9.96IFLRR92 pKa = 11.84IITEE96 pKa = 4.16DD97 pKa = 2.99GFEE100 pKa = 4.29IIRR103 pKa = 11.84RR104 pKa = 11.84YY105 pKa = 8.87STFGLFCFLEE115 pKa = 5.05RR116 pKa = 11.84ITDD119 pKa = 3.9CTSSITDD126 pKa = 4.97LIAIANSWHH135 pKa = 6.53HH136 pKa = 5.97SLNNRR141 pKa = 11.84GFNIPLADD149 pKa = 3.68VPFEE153 pKa = 4.15PVGGPDD159 pKa = 4.18DD160 pKa = 4.34DD161 pKa = 4.09QGPGGGGPDD170 pKa = 4.97DD171 pKa = 6.5DD172 pKa = 6.97DD173 pKa = 7.49DD174 pKa = 6.4DD175 pKa = 5.01DD176 pKa = 5.27HH177 pKa = 7.07GPPGDD182 pKa = 4.1VADD185 pKa = 5.21QPPQPPGDD193 pKa = 4.49DD194 pKa = 4.24DD195 pKa = 5.48ADD197 pKa = 4.04SDD199 pKa = 3.92EE200 pKa = 5.4HH201 pKa = 8.19SDD203 pKa = 3.52NGGDD207 pKa = 3.61NGEE210 pKa = 4.59GGAPQDD216 pKa = 3.97PNQIVQDD223 pKa = 4.06NNLEE227 pKa = 4.5AGPQDD232 pKa = 5.32DD233 pKa = 4.2IPILIEE239 pKa = 4.13PDD241 pKa = 3.26HH242 pKa = 7.05PGGQDD247 pKa = 2.72AVQQVVEE254 pKa = 5.03IGPPSNSPYY263 pKa = 10.47DD264 pKa = 3.59YY265 pKa = 10.54PYY267 pKa = 11.27FNGFDD272 pKa = 3.66YY273 pKa = 11.29LGFDD277 pKa = 4.53PEE279 pKa = 4.28GLNLGEE285 pKa = 4.9NEE287 pKa = 4.67LDD289 pKa = 4.1NPALNQPILQDD300 pKa = 4.59DD301 pKa = 4.57LEE303 pKa = 4.33QLLGEE308 pKa = 4.59AVVEE312 pKa = 4.95DD313 pKa = 5.12DD314 pKa = 5.05FDD316 pKa = 6.05DD317 pKa = 4.78PFDD320 pKa = 4.4VNIIDD325 pKa = 4.17VNQNNQLVPGDD336 pKa = 4.07SEE338 pKa = 4.84QPDD341 pKa = 3.71SEE343 pKa = 4.48ANEE346 pKa = 3.9NGFYY350 pKa = 11.04DD351 pKa = 4.67FDD353 pKa = 4.16VEE355 pKa = 4.26NN356 pKa = 4.08
MM1 pKa = 7.29NLIITLFAILIHH13 pKa = 6.65VSICLDD19 pKa = 3.47PVDD22 pKa = 5.53IEE24 pKa = 4.5LLQPLPEE31 pKa = 4.22EE32 pKa = 5.07GIVSEE37 pKa = 4.44EE38 pKa = 4.13VTFHH42 pKa = 6.89GIHH45 pKa = 5.53YY46 pKa = 9.47QIIVPLGQYY55 pKa = 7.65YY56 pKa = 9.95FRR58 pKa = 11.84TISYY62 pKa = 9.66NNNPIFQEE70 pKa = 4.0AQRR73 pKa = 11.84ACLKK77 pKa = 10.56VVIVTSDD84 pKa = 3.3HH85 pKa = 5.96IKK87 pKa = 10.87YY88 pKa = 9.96IFLRR92 pKa = 11.84IITEE96 pKa = 4.16DD97 pKa = 2.99GFEE100 pKa = 4.29IIRR103 pKa = 11.84RR104 pKa = 11.84YY105 pKa = 8.87STFGLFCFLEE115 pKa = 5.05RR116 pKa = 11.84ITDD119 pKa = 3.9CTSSITDD126 pKa = 4.97LIAIANSWHH135 pKa = 6.53HH136 pKa = 5.97SLNNRR141 pKa = 11.84GFNIPLADD149 pKa = 3.68VPFEE153 pKa = 4.15PVGGPDD159 pKa = 4.18DD160 pKa = 4.34DD161 pKa = 4.09QGPGGGGPDD170 pKa = 4.97DD171 pKa = 6.5DD172 pKa = 6.97DD173 pKa = 7.49DD174 pKa = 6.4DD175 pKa = 5.01DD176 pKa = 5.27HH177 pKa = 7.07GPPGDD182 pKa = 4.1VADD185 pKa = 5.21QPPQPPGDD193 pKa = 4.49DD194 pKa = 4.24DD195 pKa = 5.48ADD197 pKa = 4.04SDD199 pKa = 3.92EE200 pKa = 5.4HH201 pKa = 8.19SDD203 pKa = 3.52NGGDD207 pKa = 3.61NGEE210 pKa = 4.59GGAPQDD216 pKa = 3.97PNQIVQDD223 pKa = 4.06NNLEE227 pKa = 4.5AGPQDD232 pKa = 5.32DD233 pKa = 4.2IPILIEE239 pKa = 4.13PDD241 pKa = 3.26HH242 pKa = 7.05PGGQDD247 pKa = 2.72AVQQVVEE254 pKa = 5.03IGPPSNSPYY263 pKa = 10.47DD264 pKa = 3.59YY265 pKa = 10.54PYY267 pKa = 11.27FNGFDD272 pKa = 3.66YY273 pKa = 11.29LGFDD277 pKa = 4.53PEE279 pKa = 4.28GLNLGEE285 pKa = 4.9NEE287 pKa = 4.67LDD289 pKa = 4.1NPALNQPILQDD300 pKa = 4.59DD301 pKa = 4.57LEE303 pKa = 4.33QLLGEE308 pKa = 4.59AVVEE312 pKa = 4.95DD313 pKa = 5.12DD314 pKa = 5.05FDD316 pKa = 6.05DD317 pKa = 4.78PFDD320 pKa = 4.4VNIIDD325 pKa = 4.17VNQNNQLVPGDD336 pKa = 4.07SEE338 pKa = 4.84QPDD341 pKa = 3.71SEE343 pKa = 4.48ANEE346 pKa = 3.9NGFYY350 pKa = 11.04DD351 pKa = 4.67FDD353 pKa = 4.16VEE355 pKa = 4.26NN356 pKa = 4.08
Molecular weight: 39.19 kDa
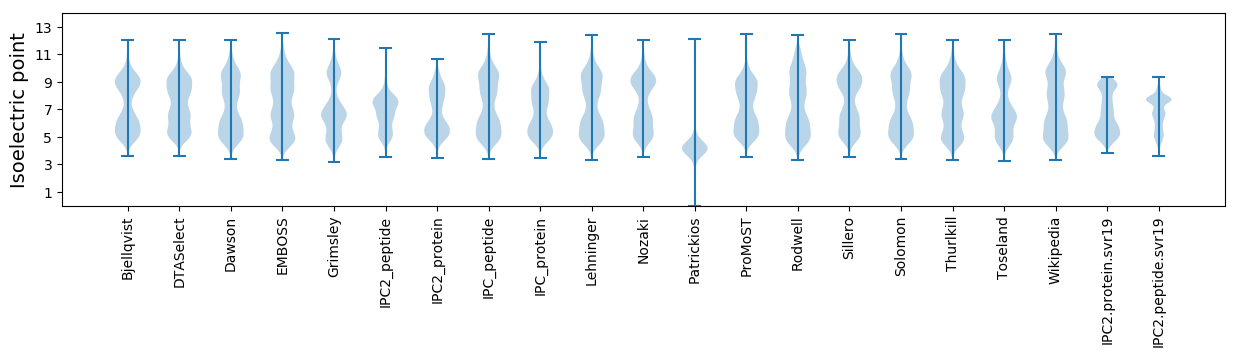
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4UCH8|Q4UCH8_THEAN Uncharacterized protein OS=Theileria annulata OX=5874 GN=TA03600 PE=4 SV=1
MM1 pKa = 7.46FKK3 pKa = 10.63YY4 pKa = 10.74LSQRR8 pKa = 11.84FFSSVYY14 pKa = 10.87ANLKK18 pKa = 9.78RR19 pKa = 11.84IRR21 pKa = 11.84PEE23 pKa = 3.75YY24 pKa = 11.12VDD26 pKa = 5.38FEE28 pKa = 5.26TNFPKK33 pKa = 10.82VKK35 pKa = 9.64IEE37 pKa = 3.97RR38 pKa = 11.84QLLPVPYY45 pKa = 8.45TQVRR49 pKa = 11.84ILFFYY54 pKa = 10.67NEE56 pKa = 3.81KK57 pKa = 10.54NLRR60 pKa = 11.84FLKK63 pKa = 10.64NLEE66 pKa = 4.25TPQTLSNCRR75 pKa = 11.84VVEE78 pKa = 3.84QLSIRR83 pKa = 11.84RR84 pKa = 11.84VKK86 pKa = 10.72FGGRR90 pKa = 11.84NNTGRR95 pKa = 11.84ITTRR99 pKa = 11.84HH100 pKa = 5.88RR101 pKa = 11.84GGGHH105 pKa = 4.21VQRR108 pKa = 11.84IRR110 pKa = 11.84LIDD113 pKa = 3.92FKK115 pKa = 10.73RR116 pKa = 11.84QRR118 pKa = 11.84KK119 pKa = 9.77DD120 pKa = 2.67IFSTVLRR127 pKa = 11.84IEE129 pKa = 4.14YY130 pKa = 10.42DD131 pKa = 3.53PTRR134 pKa = 11.84SAHH137 pKa = 5.51VALIQYY143 pKa = 10.78DD144 pKa = 4.26DD145 pKa = 4.31GVLSYY150 pKa = 9.95ILCAAGVLPGTRR162 pKa = 11.84LLSSSNAPLLPGNSLPLRR180 pKa = 11.84HH181 pKa = 6.43IPVNSIVHH189 pKa = 5.19NVEE192 pKa = 3.66MRR194 pKa = 11.84PGAGGQIARR203 pKa = 11.84SGGVYY208 pKa = 9.06CTVTEE213 pKa = 4.07KK214 pKa = 11.32DD215 pKa = 3.13EE216 pKa = 4.36TFATLRR222 pKa = 11.84LSSTEE227 pKa = 3.74LRR229 pKa = 11.84KK230 pKa = 9.84FPLDD234 pKa = 3.1CWATIGQISNIKK246 pKa = 9.77HH247 pKa = 5.61NEE249 pKa = 4.07RR250 pKa = 11.84ILKK253 pKa = 9.99KK254 pKa = 10.61AGTRR258 pKa = 11.84RR259 pKa = 11.84NMGWRR264 pKa = 11.84PVVRR268 pKa = 11.84GIAMNANSHH277 pKa = 5.37PHH279 pKa = 6.39GGGNDD284 pKa = 3.14KK285 pKa = 11.22SGTKK289 pKa = 9.77RR290 pKa = 11.84PKK292 pKa = 9.52CSIWGICKK300 pKa = 10.28DD301 pKa = 3.66GYY303 pKa = 8.72KK304 pKa = 9.58TRR306 pKa = 11.84SKK308 pKa = 10.56HH309 pKa = 5.68KK310 pKa = 9.14PLGFIIRR317 pKa = 11.84RR318 pKa = 11.84KK319 pKa = 9.51LCGRR323 pKa = 11.84LMKK326 pKa = 10.44KK327 pKa = 10.47YY328 pKa = 10.52GITKK332 pKa = 10.22RR333 pKa = 11.84PP334 pKa = 3.26
MM1 pKa = 7.46FKK3 pKa = 10.63YY4 pKa = 10.74LSQRR8 pKa = 11.84FFSSVYY14 pKa = 10.87ANLKK18 pKa = 9.78RR19 pKa = 11.84IRR21 pKa = 11.84PEE23 pKa = 3.75YY24 pKa = 11.12VDD26 pKa = 5.38FEE28 pKa = 5.26TNFPKK33 pKa = 10.82VKK35 pKa = 9.64IEE37 pKa = 3.97RR38 pKa = 11.84QLLPVPYY45 pKa = 8.45TQVRR49 pKa = 11.84ILFFYY54 pKa = 10.67NEE56 pKa = 3.81KK57 pKa = 10.54NLRR60 pKa = 11.84FLKK63 pKa = 10.64NLEE66 pKa = 4.25TPQTLSNCRR75 pKa = 11.84VVEE78 pKa = 3.84QLSIRR83 pKa = 11.84RR84 pKa = 11.84VKK86 pKa = 10.72FGGRR90 pKa = 11.84NNTGRR95 pKa = 11.84ITTRR99 pKa = 11.84HH100 pKa = 5.88RR101 pKa = 11.84GGGHH105 pKa = 4.21VQRR108 pKa = 11.84IRR110 pKa = 11.84LIDD113 pKa = 3.92FKK115 pKa = 10.73RR116 pKa = 11.84QRR118 pKa = 11.84KK119 pKa = 9.77DD120 pKa = 2.67IFSTVLRR127 pKa = 11.84IEE129 pKa = 4.14YY130 pKa = 10.42DD131 pKa = 3.53PTRR134 pKa = 11.84SAHH137 pKa = 5.51VALIQYY143 pKa = 10.78DD144 pKa = 4.26DD145 pKa = 4.31GVLSYY150 pKa = 9.95ILCAAGVLPGTRR162 pKa = 11.84LLSSSNAPLLPGNSLPLRR180 pKa = 11.84HH181 pKa = 6.43IPVNSIVHH189 pKa = 5.19NVEE192 pKa = 3.66MRR194 pKa = 11.84PGAGGQIARR203 pKa = 11.84SGGVYY208 pKa = 9.06CTVTEE213 pKa = 4.07KK214 pKa = 11.32DD215 pKa = 3.13EE216 pKa = 4.36TFATLRR222 pKa = 11.84LSSTEE227 pKa = 3.74LRR229 pKa = 11.84KK230 pKa = 9.84FPLDD234 pKa = 3.1CWATIGQISNIKK246 pKa = 9.77HH247 pKa = 5.61NEE249 pKa = 4.07RR250 pKa = 11.84ILKK253 pKa = 9.99KK254 pKa = 10.61AGTRR258 pKa = 11.84RR259 pKa = 11.84NMGWRR264 pKa = 11.84PVVRR268 pKa = 11.84GIAMNANSHH277 pKa = 5.37PHH279 pKa = 6.39GGGNDD284 pKa = 3.14KK285 pKa = 11.22SGTKK289 pKa = 9.77RR290 pKa = 11.84PKK292 pKa = 9.52CSIWGICKK300 pKa = 10.28DD301 pKa = 3.66GYY303 pKa = 8.72KK304 pKa = 9.58TRR306 pKa = 11.84SKK308 pKa = 10.56HH309 pKa = 5.68KK310 pKa = 9.14PLGFIIRR317 pKa = 11.84RR318 pKa = 11.84KK319 pKa = 9.51LCGRR323 pKa = 11.84LMKK326 pKa = 10.44KK327 pKa = 10.47YY328 pKa = 10.52GITKK332 pKa = 10.22RR333 pKa = 11.84PP334 pKa = 3.26
Molecular weight: 38.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2024948 |
38 |
5808 |
534.3 |
61.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.338 ± 0.039 | 1.648 ± 0.02 |
5.765 ± 0.032 | 6.484 ± 0.043 |
4.962 ± 0.026 | 4.487 ± 0.035 |
2.021 ± 0.012 | 7.118 ± 0.051 |
8.059 ± 0.035 | 10.089 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.054 ± 0.015 | 7.634 ± 0.054 |
3.716 ± 0.037 | 3.078 ± 0.028 |
3.827 ± 0.031 | 8.631 ± 0.038 |
6.011 ± 0.045 | 5.91 ± 0.029 |
0.814 ± 0.01 | 4.354 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
