
Immundisolibacter cernigliae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Immundisolibacterales; Immundisolibacteraceae; Immundisolibacter
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
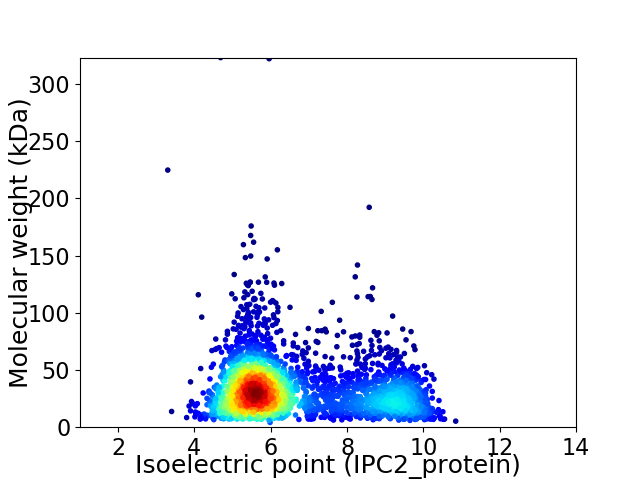
Virtual 2D-PAGE plot for 2904 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B1YSF9|A0A1B1YSF9_9GAMM Uncharacterized protein OS=Immundisolibacter cernigliae OX=1810504 GN=PG2T_05295 PE=4 SV=1
MM1 pKa = 7.41LAATASASLLATDD14 pKa = 4.78AASAGAKK21 pKa = 10.06FSIDD25 pKa = 3.23DD26 pKa = 3.86THH28 pKa = 6.12WVSVGAGLRR37 pKa = 11.84TSFSSVEE44 pKa = 3.98DD45 pKa = 3.59AAPDD49 pKa = 3.79DD50 pKa = 4.07GRR52 pKa = 11.84SNDD55 pKa = 3.46FTVDD59 pKa = 4.2SIRR62 pKa = 11.84LYY64 pKa = 11.4LNAQIHH70 pKa = 5.12QYY72 pKa = 10.57ILLEE76 pKa = 4.42FNTEE80 pKa = 4.06RR81 pKa = 11.84YY82 pKa = 7.8DD83 pKa = 3.87TGDD86 pKa = 3.77DD87 pKa = 3.5DD88 pKa = 6.67DD89 pKa = 5.83IRR91 pKa = 11.84MLDD94 pKa = 4.36AIAKK98 pKa = 7.18FTIAPEE104 pKa = 3.84FNIWAGRR111 pKa = 11.84HH112 pKa = 5.53LPPSDD117 pKa = 3.34RR118 pKa = 11.84ANLDD122 pKa = 2.99GPYY125 pKa = 10.47YY126 pKa = 10.5LNAWTFPIAQAYY138 pKa = 6.76PAIFAGRR145 pKa = 11.84DD146 pKa = 3.08EE147 pKa = 4.54GVSVNGSIDD156 pKa = 3.43EE157 pKa = 4.45GVFGYY162 pKa = 11.27ALGVYY167 pKa = 10.27DD168 pKa = 6.17GMDD171 pKa = 3.05ASYY174 pKa = 11.37SGITDD179 pKa = 4.47LNNPNQDD186 pKa = 3.81DD187 pKa = 4.02NLLFAGRR194 pKa = 11.84LQWALWDD201 pKa = 4.34PEE203 pKa = 4.26PGFYY207 pKa = 7.65TTSSYY212 pKa = 10.87FGEE215 pKa = 4.26KK216 pKa = 10.46NILTFGAAAQYY227 pKa = 10.74QSDD230 pKa = 4.09GTGTMASPGDD240 pKa = 3.66FFGWNVDD247 pKa = 3.24ALLEE251 pKa = 4.24RR252 pKa = 11.84KK253 pKa = 9.89VGDD256 pKa = 3.48GGAVSLEE263 pKa = 3.84GAYY266 pKa = 10.57YY267 pKa = 10.4DD268 pKa = 4.28YY269 pKa = 11.8DD270 pKa = 5.11HH271 pKa = 7.76DD272 pKa = 5.44DD273 pKa = 3.48EE274 pKa = 4.96MPAFGYY280 pKa = 10.02QGNGYY285 pKa = 9.17YY286 pKa = 10.94LLASFLTPQKK296 pKa = 10.65FGIGKK301 pKa = 7.99LQPHH305 pKa = 5.79VRR307 pKa = 11.84YY308 pKa = 9.32QAVDD312 pKa = 3.56DD313 pKa = 5.38DD314 pKa = 4.37NAPDD318 pKa = 3.56HH319 pKa = 7.09DD320 pKa = 4.02RR321 pKa = 11.84WEE323 pKa = 4.51LGLGYY328 pKa = 10.0IIDD331 pKa = 3.95GQKK334 pKa = 10.87AKK336 pKa = 10.97VIATYY341 pKa = 10.35GADD344 pKa = 3.75DD345 pKa = 3.94FDD347 pKa = 5.88GGEE350 pKa = 4.14TTDD353 pKa = 4.64FFILGVQLQII363 pKa = 4.54
MM1 pKa = 7.41LAATASASLLATDD14 pKa = 4.78AASAGAKK21 pKa = 10.06FSIDD25 pKa = 3.23DD26 pKa = 3.86THH28 pKa = 6.12WVSVGAGLRR37 pKa = 11.84TSFSSVEE44 pKa = 3.98DD45 pKa = 3.59AAPDD49 pKa = 3.79DD50 pKa = 4.07GRR52 pKa = 11.84SNDD55 pKa = 3.46FTVDD59 pKa = 4.2SIRR62 pKa = 11.84LYY64 pKa = 11.4LNAQIHH70 pKa = 5.12QYY72 pKa = 10.57ILLEE76 pKa = 4.42FNTEE80 pKa = 4.06RR81 pKa = 11.84YY82 pKa = 7.8DD83 pKa = 3.87TGDD86 pKa = 3.77DD87 pKa = 3.5DD88 pKa = 6.67DD89 pKa = 5.83IRR91 pKa = 11.84MLDD94 pKa = 4.36AIAKK98 pKa = 7.18FTIAPEE104 pKa = 3.84FNIWAGRR111 pKa = 11.84HH112 pKa = 5.53LPPSDD117 pKa = 3.34RR118 pKa = 11.84ANLDD122 pKa = 2.99GPYY125 pKa = 10.47YY126 pKa = 10.5LNAWTFPIAQAYY138 pKa = 6.76PAIFAGRR145 pKa = 11.84DD146 pKa = 3.08EE147 pKa = 4.54GVSVNGSIDD156 pKa = 3.43EE157 pKa = 4.45GVFGYY162 pKa = 11.27ALGVYY167 pKa = 10.27DD168 pKa = 6.17GMDD171 pKa = 3.05ASYY174 pKa = 11.37SGITDD179 pKa = 4.47LNNPNQDD186 pKa = 3.81DD187 pKa = 4.02NLLFAGRR194 pKa = 11.84LQWALWDD201 pKa = 4.34PEE203 pKa = 4.26PGFYY207 pKa = 7.65TTSSYY212 pKa = 10.87FGEE215 pKa = 4.26KK216 pKa = 10.46NILTFGAAAQYY227 pKa = 10.74QSDD230 pKa = 4.09GTGTMASPGDD240 pKa = 3.66FFGWNVDD247 pKa = 3.24ALLEE251 pKa = 4.24RR252 pKa = 11.84KK253 pKa = 9.89VGDD256 pKa = 3.48GGAVSLEE263 pKa = 3.84GAYY266 pKa = 10.57YY267 pKa = 10.4DD268 pKa = 4.28YY269 pKa = 11.8DD270 pKa = 5.11HH271 pKa = 7.76DD272 pKa = 5.44DD273 pKa = 3.48EE274 pKa = 4.96MPAFGYY280 pKa = 10.02QGNGYY285 pKa = 9.17YY286 pKa = 10.94LLASFLTPQKK296 pKa = 10.65FGIGKK301 pKa = 7.99LQPHH305 pKa = 5.79VRR307 pKa = 11.84YY308 pKa = 9.32QAVDD312 pKa = 3.56DD313 pKa = 5.38DD314 pKa = 4.37NAPDD318 pKa = 3.56HH319 pKa = 7.09DD320 pKa = 4.02RR321 pKa = 11.84WEE323 pKa = 4.51LGLGYY328 pKa = 10.0IIDD331 pKa = 3.95GQKK334 pKa = 10.87AKK336 pKa = 10.97VIATYY341 pKa = 10.35GADD344 pKa = 3.75DD345 pKa = 3.94FDD347 pKa = 5.88GGEE350 pKa = 4.14TTDD353 pKa = 4.64FFILGVQLQII363 pKa = 4.54
Molecular weight: 39.61 kDa
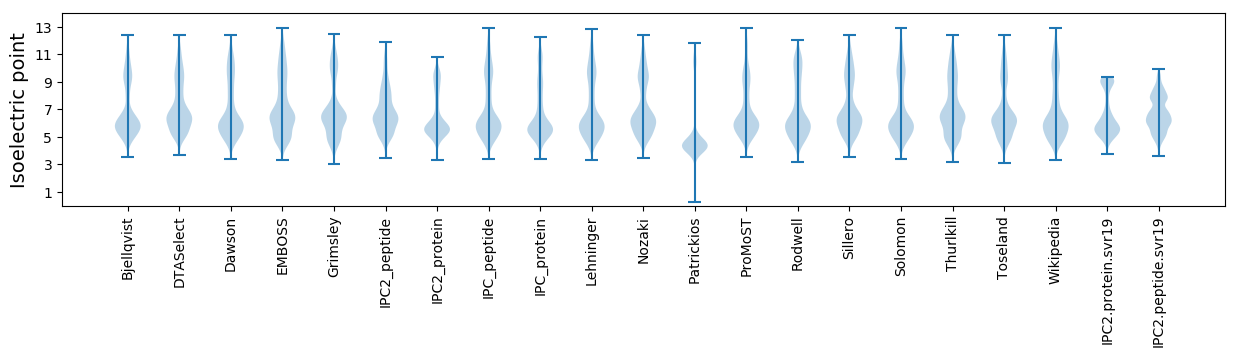
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B1YWA4|A0A1B1YWA4_9GAMM EF-P post-translational modification enzyme B OS=Immundisolibacter cernigliae OX=1810504 GN=PG2T_12415 PE=3 SV=1
MM1 pKa = 7.59PRR3 pKa = 11.84TRR5 pKa = 11.84ILTAIVLLGLVLLLLWRR22 pKa = 11.84GTLVWLLGVFAAVALRR38 pKa = 11.84AAWEE42 pKa = 3.99WAGLCSGLSGARR54 pKa = 11.84RR55 pKa = 11.84WLLLATLGASLAGAYY70 pKa = 9.1ALPASWTAVALAAGLAWWALGLRR93 pKa = 11.84LVLCYY98 pKa = 9.6PVLPDD103 pKa = 4.13WIDD106 pKa = 3.43RR107 pKa = 11.84PLAQAAIILLALAPAWLALARR128 pKa = 11.84LADD131 pKa = 3.89GQRR134 pKa = 11.84PLLLLCLALVWAADD148 pKa = 3.3SGAYY152 pKa = 9.95LVGRR156 pKa = 11.84RR157 pKa = 11.84FGRR160 pKa = 11.84RR161 pKa = 11.84KK162 pKa = 9.88LCPQVSPGKK171 pKa = 8.44TVEE174 pKa = 4.08GLLGGLVGAALIGLVAGLALGLDD197 pKa = 4.3GLRR200 pKa = 11.84LAALVGLATACAAVSVVGDD219 pKa = 3.62LAEE222 pKa = 4.35SLFKK226 pKa = 10.75RR227 pKa = 11.84RR228 pKa = 11.84AGVKK232 pKa = 10.0DD233 pKa = 3.7SSHH236 pKa = 6.94LLPGHH241 pKa = 6.33GGVLDD246 pKa = 5.45RR247 pKa = 11.84IDD249 pKa = 4.28ALIAAAPLFALTAPLLVRR267 pKa = 11.84GG268 pKa = 4.23
MM1 pKa = 7.59PRR3 pKa = 11.84TRR5 pKa = 11.84ILTAIVLLGLVLLLLWRR22 pKa = 11.84GTLVWLLGVFAAVALRR38 pKa = 11.84AAWEE42 pKa = 3.99WAGLCSGLSGARR54 pKa = 11.84RR55 pKa = 11.84WLLLATLGASLAGAYY70 pKa = 9.1ALPASWTAVALAAGLAWWALGLRR93 pKa = 11.84LVLCYY98 pKa = 9.6PVLPDD103 pKa = 4.13WIDD106 pKa = 3.43RR107 pKa = 11.84PLAQAAIILLALAPAWLALARR128 pKa = 11.84LADD131 pKa = 3.89GQRR134 pKa = 11.84PLLLLCLALVWAADD148 pKa = 3.3SGAYY152 pKa = 9.95LVGRR156 pKa = 11.84RR157 pKa = 11.84FGRR160 pKa = 11.84RR161 pKa = 11.84KK162 pKa = 9.88LCPQVSPGKK171 pKa = 8.44TVEE174 pKa = 4.08GLLGGLVGAALIGLVAGLALGLDD197 pKa = 4.3GLRR200 pKa = 11.84LAALVGLATACAAVSVVGDD219 pKa = 3.62LAEE222 pKa = 4.35SLFKK226 pKa = 10.75RR227 pKa = 11.84RR228 pKa = 11.84AGVKK232 pKa = 10.0DD233 pKa = 3.7SSHH236 pKa = 6.94LLPGHH241 pKa = 6.33GGVLDD246 pKa = 5.45RR247 pKa = 11.84IDD249 pKa = 4.28ALIAAAPLFALTAPLLVRR267 pKa = 11.84GG268 pKa = 4.23
Molecular weight: 27.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932638 |
41 |
3242 |
321.2 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.257 ± 0.074 | 0.988 ± 0.018 |
5.754 ± 0.033 | 5.209 ± 0.039 |
3.35 ± 0.029 | 8.823 ± 0.046 |
2.368 ± 0.026 | 4.045 ± 0.029 |
2.315 ± 0.032 | 11.582 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.022 | 2.239 ± 0.025 |
5.583 ± 0.035 | 3.857 ± 0.028 |
7.877 ± 0.044 | 4.451 ± 0.033 |
4.83 ± 0.039 | 7.552 ± 0.038 |
1.427 ± 0.022 | 2.418 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
