
Mesotoga sp. SC_3PWM13N19
Taxonomy: cellular organisms; Bacteria; Thermotogae; Thermotogae; Kosmotogales; Kosmotogaceae; Mesotoga; unclassified Mesotoga
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
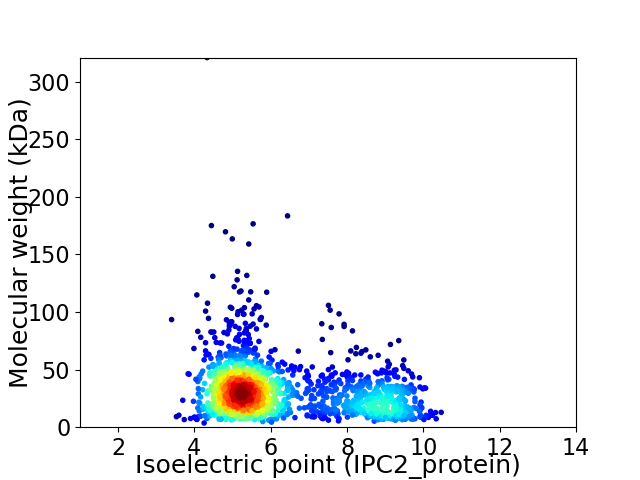
Virtual 2D-PAGE plot for 1662 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328IK35|A0A328IK35_9BACT Uncharacterized protein OS=Mesotoga sp. SC_3PWM13N19 OX=1487951 GN=DS66_06215 PE=4 SV=1
MM1 pKa = 7.2KK2 pKa = 10.25RR3 pKa = 11.84AIGFVAIGLLVLVIAGCFSLDD24 pKa = 3.36QKK26 pKa = 11.13AVNSEE31 pKa = 4.06EE32 pKa = 3.91VSFILNDD39 pKa = 4.16DD40 pKa = 3.65GSISIKK46 pKa = 10.39ALKK49 pKa = 9.63EE50 pKa = 3.55LTGIDD55 pKa = 3.86VIVAASIEE63 pKa = 4.22EE64 pKa = 3.99EE65 pKa = 4.87AITVDD70 pKa = 4.81DD71 pKa = 4.32SLLKK75 pKa = 10.34IVTHH79 pKa = 6.18SDD81 pKa = 2.78EE82 pKa = 4.27GTTRR86 pKa = 11.84IAIVSTSANIRR97 pKa = 11.84EE98 pKa = 4.36GEE100 pKa = 4.14EE101 pKa = 3.55IARR104 pKa = 11.84IDD106 pKa = 4.6GNFTTLKK113 pKa = 10.02NLVISSSAKK122 pKa = 8.5YY123 pKa = 10.9LEE125 pKa = 4.38MPSPKK130 pKa = 9.05TQAKK134 pKa = 9.73PPFPDD139 pKa = 5.06GISIVDD145 pKa = 3.64GSIDD149 pKa = 3.55KK150 pKa = 10.95ASNGYY155 pKa = 9.64FLVHH159 pKa = 6.7AEE161 pKa = 4.21NVNGFAGVEE170 pKa = 3.71ITITYY175 pKa = 9.13DD176 pKa = 3.11PQYY179 pKa = 11.02IIIDD183 pKa = 3.75KK184 pKa = 10.97SKK186 pKa = 10.7GEE188 pKa = 4.0QGVTPLNSFGTGLLIVQHH206 pKa = 5.9TEE208 pKa = 3.35NTITITSAFNSAKK221 pKa = 10.29SVNCEE226 pKa = 3.78YY227 pKa = 10.17IYY229 pKa = 10.23QIHH232 pKa = 5.98FVASLIEE239 pKa = 4.31GKK241 pKa = 7.71TLIGLSGEE249 pKa = 4.13VRR251 pKa = 11.84DD252 pKa = 4.63ANTGLIQTVFHH263 pKa = 7.28DD264 pKa = 4.33GEE266 pKa = 4.28ITIGGPKK273 pKa = 10.37LIGDD277 pKa = 4.55FDD279 pKa = 4.15NSNKK283 pKa = 9.39VDD285 pKa = 4.06LPDD288 pKa = 5.66FILFARR294 pKa = 11.84SYY296 pKa = 10.44GSVLGGEE303 pKa = 4.48GEE305 pKa = 4.4YY306 pKa = 10.34IEE308 pKa = 5.18TYY310 pKa = 10.43DD311 pKa = 4.6IAPAEE316 pKa = 4.32DD317 pKa = 4.22KK318 pKa = 11.27YY319 pKa = 10.88QGVWAGIYY327 pKa = 9.74DD328 pKa = 3.71SCATDD333 pKa = 3.98GEE335 pKa = 4.31IDD337 pKa = 3.89LVDD340 pKa = 4.93FIIFARR346 pKa = 11.84NYY348 pKa = 10.17GKK350 pKa = 10.04NKK352 pKa = 9.36PNEE355 pKa = 4.04PPFISVPDD363 pKa = 3.59QQVAQGDD370 pKa = 4.17TLEE373 pKa = 5.44LDD375 pKa = 3.8LLNYY379 pKa = 10.61SSDD382 pKa = 3.88PEE384 pKa = 5.14GDD386 pKa = 3.35SLLFILEE393 pKa = 3.96VDD395 pKa = 3.88SPGTITDD402 pKa = 4.17SIYY405 pKa = 10.7SYY407 pKa = 11.04TPDD410 pKa = 4.4FEE412 pKa = 4.79TLGNQTVDD420 pKa = 2.8IAVEE424 pKa = 4.07DD425 pKa = 3.84TAGNMMTDD433 pKa = 3.15TFIIEE438 pKa = 4.08VTKK441 pKa = 10.08TNRR444 pKa = 11.84PPVTDD449 pKa = 5.7GIPDD453 pKa = 3.28QTIAEE458 pKa = 4.48GEE460 pKa = 4.35TLNLEE465 pKa = 4.13LLDD468 pKa = 4.06YY469 pKa = 11.32FEE471 pKa = 6.61DD472 pKa = 4.68PDD474 pKa = 4.47GDD476 pKa = 3.81NLVFAVISGVGTVIGSEE493 pKa = 4.11YY494 pKa = 9.98TYY496 pKa = 11.23SPNYY500 pKa = 9.52DD501 pKa = 2.98AAGTYY506 pKa = 9.53QVTIKK511 pKa = 10.75AADD514 pKa = 3.7GNGGEE519 pKa = 4.35VEE521 pKa = 4.45TTFTLTVEE529 pKa = 4.27NTNRR533 pKa = 11.84PPEE536 pKa = 4.25EE537 pKa = 4.13PQLVSPEE544 pKa = 3.81NAALLINTTSVEE556 pKa = 4.1LSWNCSDD563 pKa = 5.7PDD565 pKa = 4.08GQLLTYY571 pKa = 10.33DD572 pKa = 3.77VYY574 pKa = 11.08FGEE577 pKa = 5.34DD578 pKa = 3.0IEE580 pKa = 4.28ALEE583 pKa = 4.69VIATQSATSLHH594 pKa = 5.22VQEE597 pKa = 4.3ITAGKK602 pKa = 7.8TYY604 pKa = 10.3FWMIVARR611 pKa = 11.84DD612 pKa = 3.45PHH614 pKa = 6.51GAEE617 pKa = 4.17TASEE621 pKa = 4.7VYY623 pKa = 10.6SFSTDD628 pKa = 2.97YY629 pKa = 11.32YY630 pKa = 10.03LIPGGEE636 pKa = 4.25FEE638 pKa = 5.48GMISGYY644 pKa = 11.66ALMTKK649 pKa = 10.34ANSPYY654 pKa = 9.99IITGDD659 pKa = 3.14IVVEE663 pKa = 4.48SGAQLIIEE671 pKa = 4.64PGVEE675 pKa = 3.88VRR677 pKa = 11.84FTYY680 pKa = 10.48ISDD683 pKa = 3.87PDD685 pKa = 3.75NNGQEE690 pKa = 4.29DD691 pKa = 4.53TNAADD696 pKa = 4.56LIVYY700 pKa = 8.52GKK702 pKa = 10.5LFAEE706 pKa = 5.33GSEE709 pKa = 4.29TEE711 pKa = 4.13PLLFTSNEE719 pKa = 3.92TPALKK724 pKa = 10.49GDD726 pKa = 3.06WGGIRR731 pKa = 11.84FASSEE736 pKa = 4.15GTSKK740 pKa = 10.32ISHH743 pKa = 6.17ATIEE747 pKa = 4.15MAKK750 pKa = 10.45DD751 pKa = 4.05GVWTSSNCNIEE762 pKa = 3.74ISNCEE767 pKa = 3.21IRR769 pKa = 11.84TNIDD773 pKa = 3.11YY774 pKa = 11.02GVLLGSNSNATIVDD788 pKa = 3.83STIHH792 pKa = 6.27LNATGISNYY801 pKa = 8.35GTMIIRR807 pKa = 11.84GCTISSSGGTGIYY820 pKa = 10.45SNGVYY825 pKa = 10.01IEE827 pKa = 4.26IYY829 pKa = 7.51EE830 pKa = 4.21TLIGEE835 pKa = 4.23NSSTGITLHH844 pKa = 6.24NSSSNSRR851 pKa = 11.84VQNCHH856 pKa = 5.92FLDD859 pKa = 3.88NGSYY863 pKa = 10.63GIEE866 pKa = 4.08GYY868 pKa = 10.13RR869 pKa = 11.84FEE871 pKa = 5.69VIDD874 pKa = 4.52SSFARR879 pKa = 11.84NGNHH883 pKa = 7.33AISGSNCVVEE893 pKa = 4.89GCLFTEE899 pKa = 4.56SIKK902 pKa = 10.8GIRR905 pKa = 11.84GSSITAVNNNFEE917 pKa = 4.77GEE919 pKa = 4.43FCSSFYY925 pKa = 10.99DD926 pKa = 4.12DD927 pKa = 3.93YY928 pKa = 11.79SGVYY932 pKa = 9.5IDD934 pKa = 4.04GQALISHH941 pKa = 6.19NVFSGFATGVWLATSSEE958 pKa = 4.08VTIQDD963 pKa = 3.42NLVTGNYY970 pKa = 9.85RR971 pKa = 11.84GICFASVDD979 pKa = 4.14GQNLTCSNNNIYY991 pKa = 10.53DD992 pKa = 3.61NRR994 pKa = 11.84DD995 pKa = 2.86WNVYY999 pKa = 10.13NDD1001 pKa = 3.53TNQRR1005 pKa = 11.84VAITNSFWGTDD1016 pKa = 3.3NEE1018 pKa = 4.46STIKK1022 pKa = 10.82SKK1024 pKa = 10.75IFDD1027 pKa = 3.89YY1028 pKa = 11.61YY1029 pKa = 10.63EE1030 pKa = 4.75DD1031 pKa = 3.78SSKK1034 pKa = 11.41GPVSYY1039 pKa = 10.23TGFKK1043 pKa = 10.61SSLIDD1048 pKa = 3.45GATSSKK1054 pKa = 10.3RR1055 pKa = 11.84IALIPYY1061 pKa = 9.64DD1062 pKa = 3.52RR1063 pKa = 5.93
MM1 pKa = 7.2KK2 pKa = 10.25RR3 pKa = 11.84AIGFVAIGLLVLVIAGCFSLDD24 pKa = 3.36QKK26 pKa = 11.13AVNSEE31 pKa = 4.06EE32 pKa = 3.91VSFILNDD39 pKa = 4.16DD40 pKa = 3.65GSISIKK46 pKa = 10.39ALKK49 pKa = 9.63EE50 pKa = 3.55LTGIDD55 pKa = 3.86VIVAASIEE63 pKa = 4.22EE64 pKa = 3.99EE65 pKa = 4.87AITVDD70 pKa = 4.81DD71 pKa = 4.32SLLKK75 pKa = 10.34IVTHH79 pKa = 6.18SDD81 pKa = 2.78EE82 pKa = 4.27GTTRR86 pKa = 11.84IAIVSTSANIRR97 pKa = 11.84EE98 pKa = 4.36GEE100 pKa = 4.14EE101 pKa = 3.55IARR104 pKa = 11.84IDD106 pKa = 4.6GNFTTLKK113 pKa = 10.02NLVISSSAKK122 pKa = 8.5YY123 pKa = 10.9LEE125 pKa = 4.38MPSPKK130 pKa = 9.05TQAKK134 pKa = 9.73PPFPDD139 pKa = 5.06GISIVDD145 pKa = 3.64GSIDD149 pKa = 3.55KK150 pKa = 10.95ASNGYY155 pKa = 9.64FLVHH159 pKa = 6.7AEE161 pKa = 4.21NVNGFAGVEE170 pKa = 3.71ITITYY175 pKa = 9.13DD176 pKa = 3.11PQYY179 pKa = 11.02IIIDD183 pKa = 3.75KK184 pKa = 10.97SKK186 pKa = 10.7GEE188 pKa = 4.0QGVTPLNSFGTGLLIVQHH206 pKa = 5.9TEE208 pKa = 3.35NTITITSAFNSAKK221 pKa = 10.29SVNCEE226 pKa = 3.78YY227 pKa = 10.17IYY229 pKa = 10.23QIHH232 pKa = 5.98FVASLIEE239 pKa = 4.31GKK241 pKa = 7.71TLIGLSGEE249 pKa = 4.13VRR251 pKa = 11.84DD252 pKa = 4.63ANTGLIQTVFHH263 pKa = 7.28DD264 pKa = 4.33GEE266 pKa = 4.28ITIGGPKK273 pKa = 10.37LIGDD277 pKa = 4.55FDD279 pKa = 4.15NSNKK283 pKa = 9.39VDD285 pKa = 4.06LPDD288 pKa = 5.66FILFARR294 pKa = 11.84SYY296 pKa = 10.44GSVLGGEE303 pKa = 4.48GEE305 pKa = 4.4YY306 pKa = 10.34IEE308 pKa = 5.18TYY310 pKa = 10.43DD311 pKa = 4.6IAPAEE316 pKa = 4.32DD317 pKa = 4.22KK318 pKa = 11.27YY319 pKa = 10.88QGVWAGIYY327 pKa = 9.74DD328 pKa = 3.71SCATDD333 pKa = 3.98GEE335 pKa = 4.31IDD337 pKa = 3.89LVDD340 pKa = 4.93FIIFARR346 pKa = 11.84NYY348 pKa = 10.17GKK350 pKa = 10.04NKK352 pKa = 9.36PNEE355 pKa = 4.04PPFISVPDD363 pKa = 3.59QQVAQGDD370 pKa = 4.17TLEE373 pKa = 5.44LDD375 pKa = 3.8LLNYY379 pKa = 10.61SSDD382 pKa = 3.88PEE384 pKa = 5.14GDD386 pKa = 3.35SLLFILEE393 pKa = 3.96VDD395 pKa = 3.88SPGTITDD402 pKa = 4.17SIYY405 pKa = 10.7SYY407 pKa = 11.04TPDD410 pKa = 4.4FEE412 pKa = 4.79TLGNQTVDD420 pKa = 2.8IAVEE424 pKa = 4.07DD425 pKa = 3.84TAGNMMTDD433 pKa = 3.15TFIIEE438 pKa = 4.08VTKK441 pKa = 10.08TNRR444 pKa = 11.84PPVTDD449 pKa = 5.7GIPDD453 pKa = 3.28QTIAEE458 pKa = 4.48GEE460 pKa = 4.35TLNLEE465 pKa = 4.13LLDD468 pKa = 4.06YY469 pKa = 11.32FEE471 pKa = 6.61DD472 pKa = 4.68PDD474 pKa = 4.47GDD476 pKa = 3.81NLVFAVISGVGTVIGSEE493 pKa = 4.11YY494 pKa = 9.98TYY496 pKa = 11.23SPNYY500 pKa = 9.52DD501 pKa = 2.98AAGTYY506 pKa = 9.53QVTIKK511 pKa = 10.75AADD514 pKa = 3.7GNGGEE519 pKa = 4.35VEE521 pKa = 4.45TTFTLTVEE529 pKa = 4.27NTNRR533 pKa = 11.84PPEE536 pKa = 4.25EE537 pKa = 4.13PQLVSPEE544 pKa = 3.81NAALLINTTSVEE556 pKa = 4.1LSWNCSDD563 pKa = 5.7PDD565 pKa = 4.08GQLLTYY571 pKa = 10.33DD572 pKa = 3.77VYY574 pKa = 11.08FGEE577 pKa = 5.34DD578 pKa = 3.0IEE580 pKa = 4.28ALEE583 pKa = 4.69VIATQSATSLHH594 pKa = 5.22VQEE597 pKa = 4.3ITAGKK602 pKa = 7.8TYY604 pKa = 10.3FWMIVARR611 pKa = 11.84DD612 pKa = 3.45PHH614 pKa = 6.51GAEE617 pKa = 4.17TASEE621 pKa = 4.7VYY623 pKa = 10.6SFSTDD628 pKa = 2.97YY629 pKa = 11.32YY630 pKa = 10.03LIPGGEE636 pKa = 4.25FEE638 pKa = 5.48GMISGYY644 pKa = 11.66ALMTKK649 pKa = 10.34ANSPYY654 pKa = 9.99IITGDD659 pKa = 3.14IVVEE663 pKa = 4.48SGAQLIIEE671 pKa = 4.64PGVEE675 pKa = 3.88VRR677 pKa = 11.84FTYY680 pKa = 10.48ISDD683 pKa = 3.87PDD685 pKa = 3.75NNGQEE690 pKa = 4.29DD691 pKa = 4.53TNAADD696 pKa = 4.56LIVYY700 pKa = 8.52GKK702 pKa = 10.5LFAEE706 pKa = 5.33GSEE709 pKa = 4.29TEE711 pKa = 4.13PLLFTSNEE719 pKa = 3.92TPALKK724 pKa = 10.49GDD726 pKa = 3.06WGGIRR731 pKa = 11.84FASSEE736 pKa = 4.15GTSKK740 pKa = 10.32ISHH743 pKa = 6.17ATIEE747 pKa = 4.15MAKK750 pKa = 10.45DD751 pKa = 4.05GVWTSSNCNIEE762 pKa = 3.74ISNCEE767 pKa = 3.21IRR769 pKa = 11.84TNIDD773 pKa = 3.11YY774 pKa = 11.02GVLLGSNSNATIVDD788 pKa = 3.83STIHH792 pKa = 6.27LNATGISNYY801 pKa = 8.35GTMIIRR807 pKa = 11.84GCTISSSGGTGIYY820 pKa = 10.45SNGVYY825 pKa = 10.01IEE827 pKa = 4.26IYY829 pKa = 7.51EE830 pKa = 4.21TLIGEE835 pKa = 4.23NSSTGITLHH844 pKa = 6.24NSSSNSRR851 pKa = 11.84VQNCHH856 pKa = 5.92FLDD859 pKa = 3.88NGSYY863 pKa = 10.63GIEE866 pKa = 4.08GYY868 pKa = 10.13RR869 pKa = 11.84FEE871 pKa = 5.69VIDD874 pKa = 4.52SSFARR879 pKa = 11.84NGNHH883 pKa = 7.33AISGSNCVVEE893 pKa = 4.89GCLFTEE899 pKa = 4.56SIKK902 pKa = 10.8GIRR905 pKa = 11.84GSSITAVNNNFEE917 pKa = 4.77GEE919 pKa = 4.43FCSSFYY925 pKa = 10.99DD926 pKa = 4.12DD927 pKa = 3.93YY928 pKa = 11.79SGVYY932 pKa = 9.5IDD934 pKa = 4.04GQALISHH941 pKa = 6.19NVFSGFATGVWLATSSEE958 pKa = 4.08VTIQDD963 pKa = 3.42NLVTGNYY970 pKa = 9.85RR971 pKa = 11.84GICFASVDD979 pKa = 4.14GQNLTCSNNNIYY991 pKa = 10.53DD992 pKa = 3.61NRR994 pKa = 11.84DD995 pKa = 2.86WNVYY999 pKa = 10.13NDD1001 pKa = 3.53TNQRR1005 pKa = 11.84VAITNSFWGTDD1016 pKa = 3.3NEE1018 pKa = 4.46STIKK1022 pKa = 10.82SKK1024 pKa = 10.75IFDD1027 pKa = 3.89YY1028 pKa = 11.61YY1029 pKa = 10.63EE1030 pKa = 4.75DD1031 pKa = 3.78SSKK1034 pKa = 11.41GPVSYY1039 pKa = 10.23TGFKK1043 pKa = 10.61SSLIDD1048 pKa = 3.45GATSSKK1054 pKa = 10.3RR1055 pKa = 11.84IALIPYY1061 pKa = 9.64DD1062 pKa = 3.52RR1063 pKa = 5.93
Molecular weight: 114.83 kDa
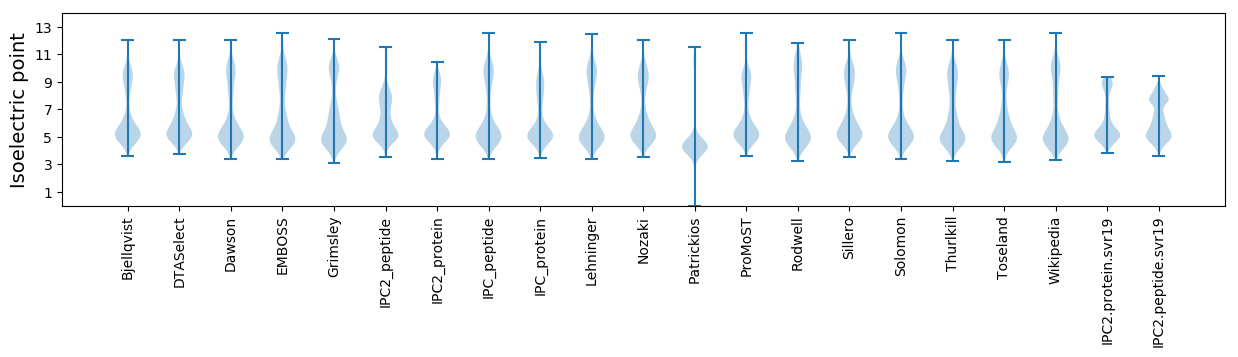
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328IT45|A0A328IT45_9BACT Sugar ABC transporter ATP-binding protein (Fragment) OS=Mesotoga sp. SC_3PWM13N19 OX=1487951 GN=DS66_03210 PE=4 SV=1
MM1 pKa = 7.17VKK3 pKa = 10.42DD4 pKa = 3.27NRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.64CRR16 pKa = 11.84LCGTKK21 pKa = 9.17TKK23 pKa = 10.96YY24 pKa = 9.91IDD26 pKa = 3.65YY27 pKa = 10.49KK28 pKa = 10.17NTSLLRR34 pKa = 11.84DD35 pKa = 3.58YY36 pKa = 10.1VTEE39 pKa = 4.11KK40 pKa = 11.08GKK42 pKa = 10.48IIPKK46 pKa = 10.17RR47 pKa = 11.84ITGNCAKK54 pKa = 9.36HH55 pKa = 4.62QRR57 pKa = 11.84MVKK60 pKa = 9.9EE61 pKa = 4.51AIQRR65 pKa = 11.84ARR67 pKa = 11.84FMALLPYY74 pKa = 9.86TKK76 pKa = 10.23EE77 pKa = 3.82
MM1 pKa = 7.17VKK3 pKa = 10.42DD4 pKa = 3.27NRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.64CRR16 pKa = 11.84LCGTKK21 pKa = 9.17TKK23 pKa = 10.96YY24 pKa = 9.91IDD26 pKa = 3.65YY27 pKa = 10.49KK28 pKa = 10.17NTSLLRR34 pKa = 11.84DD35 pKa = 3.58YY36 pKa = 10.1VTEE39 pKa = 4.11KK40 pKa = 11.08GKK42 pKa = 10.48IIPKK46 pKa = 10.17RR47 pKa = 11.84ITGNCAKK54 pKa = 9.36HH55 pKa = 4.62QRR57 pKa = 11.84MVKK60 pKa = 9.9EE61 pKa = 4.51AIQRR65 pKa = 11.84ARR67 pKa = 11.84FMALLPYY74 pKa = 9.86TKK76 pKa = 10.23EE77 pKa = 3.82
Molecular weight: 9.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
494837 |
32 |
2917 |
297.7 |
33.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.624 ± 0.057 | 0.809 ± 0.018 |
5.456 ± 0.047 | 7.729 ± 0.067 |
4.938 ± 0.052 | 7.34 ± 0.056 |
1.506 ± 0.025 | 7.511 ± 0.048 |
5.793 ± 0.057 | 10.031 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.556 ± 0.027 | 3.765 ± 0.032 |
3.7 ± 0.031 | 2.285 ± 0.026 |
5.581 ± 0.048 | 7.492 ± 0.054 |
4.903 ± 0.041 | 7.542 ± 0.049 |
1.02 ± 0.023 | 3.418 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
