
Mariniradius saccharolyticus AK6
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Mariniradius; Mariniradius saccharolyticus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
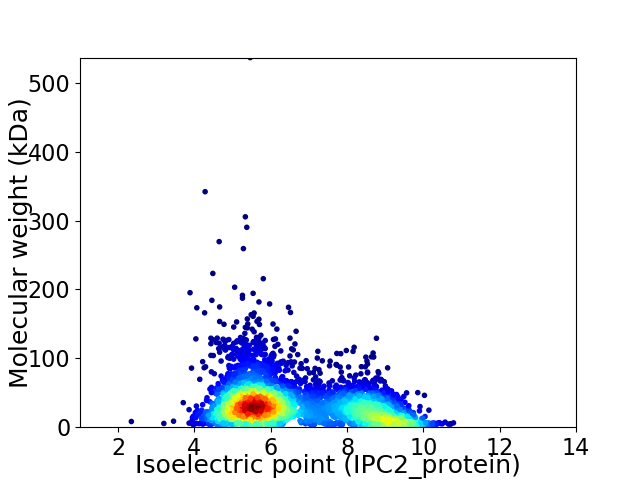
Virtual 2D-PAGE plot for 4633 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
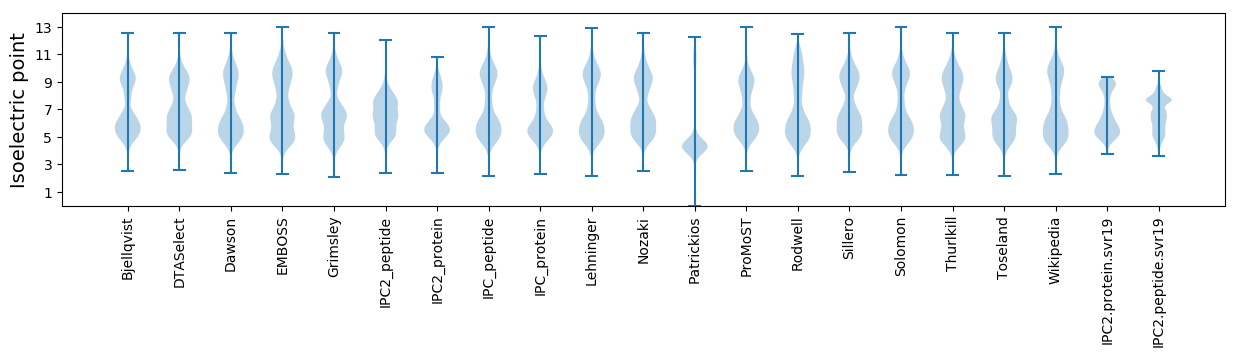
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M7X9M5|M7X9M5_9BACT Rhomboid family protein OS=Mariniradius saccharolyticus AK6 OX=1239962 GN=C943_01910 PE=4 SV=1
MM1 pKa = 7.44NPTIVFGQGFNNNEE15 pKa = 4.02WVFGDD20 pKa = 4.91CGTGQNNYY28 pKa = 10.36LSFGKK33 pKa = 10.65GGDD36 pKa = 3.82PIVRR40 pKa = 11.84TLPSGVIVGQNNNAVAIDD58 pKa = 4.74PISGNIIFYY67 pKa = 10.7SNGVLVYY74 pKa = 10.37DD75 pKa = 4.71ANNQILEE82 pKa = 4.27GVAPGINGDD91 pKa = 3.56INGQQQVGIGILSYY105 pKa = 10.54EE106 pKa = 4.13PQGDD110 pKa = 3.11RR111 pKa = 11.84LYY113 pKa = 11.06YY114 pKa = 9.84IFYY117 pKa = 10.34VSPAGDD123 pKa = 3.62LQYY126 pKa = 12.09ALIDD130 pKa = 3.72MNAPGQAVGNEE141 pKa = 4.05PPLGEE146 pKa = 5.15IIEE149 pKa = 4.36KK150 pKa = 10.49DD151 pKa = 3.5QVVVGAVNGPILVVKK166 pKa = 9.09TPQSPSYY173 pKa = 10.1LISIEE178 pKa = 3.99GGSIVSRR185 pKa = 11.84RR186 pKa = 11.84IGANPGEE193 pKa = 4.22FTLISNAAVPFTPKK207 pKa = 10.37FMVFDD212 pKa = 3.87EE213 pKa = 4.49ASQRR217 pKa = 11.84VILIPDD223 pKa = 4.32DD224 pKa = 4.12PNQNIVVLGFDD235 pKa = 3.65TASGTFGSQEE245 pKa = 4.47PISQSSNGQAYY256 pKa = 9.81GGASFSPDD264 pKa = 2.54GAFIYY269 pKa = 9.73FSRR272 pKa = 11.84GNQLFRR278 pKa = 11.84IPSNNLTASPEE289 pKa = 4.41AIPLDD294 pKa = 4.03NPVHH298 pKa = 6.93AIYY301 pKa = 10.18DD302 pKa = 3.91VKK304 pKa = 11.03VGPDD308 pKa = 2.85GRR310 pKa = 11.84LYY312 pKa = 10.67YY313 pKa = 9.94IYY315 pKa = 10.49EE316 pKa = 4.15EE317 pKa = 4.33VLGGPQLIGRR327 pKa = 11.84VNNPNEE333 pKa = 4.12PVLDD337 pKa = 4.17DD338 pKa = 4.63LVLAEE343 pKa = 5.25DD344 pKa = 4.7PFNGTDD350 pKa = 3.47FCGRR354 pKa = 11.84IFPKK358 pKa = 10.41FAPNATISPTVDD370 pKa = 4.01FTWEE374 pKa = 3.73PDD376 pKa = 3.63QPCSNNPVQLTSLITPEE393 pKa = 3.69NYY395 pKa = 9.83RR396 pKa = 11.84PVSFEE401 pKa = 3.59WTFNPPLVDD410 pKa = 3.73EE411 pKa = 5.6DD412 pKa = 4.28GQPVDD417 pKa = 3.34IDD419 pKa = 3.57FTQEE423 pKa = 3.47HH424 pKa = 6.7LLIPEE429 pKa = 4.84DD430 pKa = 3.34ATANQSIDD438 pKa = 3.25VTLTVTFADD447 pKa = 4.79GSTQTVNKK455 pKa = 9.24TISLLEE461 pKa = 4.16NDD463 pKa = 4.39LQVQFSSQDD472 pKa = 3.23TTVCEE477 pKa = 4.55GACVDD482 pKa = 3.62IGSMLEE488 pKa = 3.96VQQGGGQGGGQGGGQGNYY506 pKa = 9.45EE507 pKa = 4.24YY508 pKa = 10.0FWSNIRR514 pKa = 11.84EE515 pKa = 4.13WRR517 pKa = 11.84SSKK520 pKa = 10.77DD521 pKa = 3.24NCVDD525 pKa = 3.75LPGLYY530 pKa = 8.72WVLVRR535 pKa = 11.84EE536 pKa = 4.76PGSSCYY542 pKa = 10.37AYY544 pKa = 9.8GSVRR548 pKa = 11.84VRR550 pKa = 11.84IWDD553 pKa = 4.85LLDD556 pKa = 3.11QSNNVWYY563 pKa = 10.11FGDD566 pKa = 4.18GAGLDD571 pKa = 4.3FNPDD575 pKa = 3.63PNNPNAPLPRR585 pKa = 11.84PVGHH589 pKa = 6.26NQNIPAGTTTISDD602 pKa = 3.35GTGQVLFFTDD612 pKa = 5.3GEE614 pKa = 4.97TVWDD618 pKa = 3.94LNGNIMEE625 pKa = 4.92NGDD628 pKa = 4.23SIGGSNLSSQSVIAVPVPTDD648 pKa = 3.07EE649 pKa = 4.55TIFYY653 pKa = 10.89LFTTQLAQDD662 pKa = 4.33GSNQVKK668 pKa = 9.9FSVVDD673 pKa = 3.98IKK675 pKa = 11.45AEE677 pKa = 3.97NQNGVGNVVSKK688 pKa = 11.44DD689 pKa = 3.27NFLFSPSTEE698 pKa = 3.76HH699 pKa = 6.8SAALAAGDD707 pKa = 3.95TTWVLFHH714 pKa = 7.06EE715 pKa = 5.11LGNNTFRR722 pKa = 11.84AYY724 pKa = 9.44PVSIHH729 pKa = 7.26GIGQPVLSSVGSSHH743 pKa = 6.38GFNSGVGSMKK753 pKa = 10.43FSPDD757 pKa = 2.97GTKK760 pKa = 10.44VAVTISDD767 pKa = 4.35GACSRR772 pKa = 11.84LEE774 pKa = 4.37IFDD777 pKa = 4.46FDD779 pKa = 4.15QNTGEE784 pKa = 4.09MTEE787 pKa = 4.12YY788 pKa = 11.42ALLDD792 pKa = 3.97LGCNNDD798 pKa = 3.11QVYY801 pKa = 10.72GLEE804 pKa = 3.99FSNDD808 pKa = 2.81GSRR811 pKa = 11.84VYY813 pKa = 10.81VSYY816 pKa = 10.92RR817 pKa = 11.84GGNGKK822 pKa = 8.93IDD824 pKa = 3.76EE825 pKa = 4.75FLIQAPEE832 pKa = 3.86QTGNNPIPCATCFSSATTRR851 pKa = 11.84PQRR854 pKa = 11.84EE855 pKa = 3.86ACILNSPVRR864 pKa = 11.84NTLTTSGPFGALQIGPDD881 pKa = 3.55GQIYY885 pKa = 8.59VARR888 pKa = 11.84PGQNVLGTINAGQDD902 pKa = 4.0CNRR905 pKa = 11.84SAYY908 pKa = 10.04NEE910 pKa = 4.32NGTSTLLGTSNLGLPSFVQQSGSSIPEE937 pKa = 3.75PALAGTPFLCIDD949 pKa = 4.06PQNGALGEE957 pKa = 4.19FEE959 pKa = 5.44GGGEE963 pKa = 4.17PDD965 pKa = 2.93IDD967 pKa = 3.99SYY969 pKa = 11.62FWTIVHH975 pKa = 6.9EE976 pKa = 5.02DD977 pKa = 3.29GTSILNNYY985 pKa = 8.14GGPGEE990 pKa = 4.03NFQVLEE996 pKa = 4.2QPFTRR1001 pKa = 11.84PGTYY1005 pKa = 8.87TVSLRR1010 pKa = 11.84VDD1012 pKa = 3.06RR1013 pKa = 11.84CGDD1016 pKa = 3.37PDD1018 pKa = 4.01FYY1020 pKa = 11.05EE1021 pKa = 4.66ASLEE1025 pKa = 4.27VEE1027 pKa = 4.88VIAPPTLTLPGEE1039 pKa = 4.3ATLCSGNPVSLTAIDD1054 pKa = 5.26GYY1056 pKa = 11.51DD1057 pKa = 3.76PADD1060 pKa = 3.52GLYY1063 pKa = 10.83DD1064 pKa = 3.84FEE1066 pKa = 4.45WRR1068 pKa = 11.84NAQGTLFGDD1077 pKa = 4.11SNSNTIFVNEE1087 pKa = 3.94EE1088 pKa = 4.07SIFTVTVSHH1097 pKa = 7.0RR1098 pKa = 11.84RR1099 pKa = 11.84PDD1101 pKa = 4.52DD1102 pKa = 3.37IPAEE1106 pKa = 4.11FFNPCPATASVFVGPPFDD1124 pKa = 5.08FDD1126 pKa = 3.71LTQTADD1132 pKa = 3.26QVCFDD1137 pKa = 3.76EE1138 pKa = 4.72SLVVFAPNTPIVGEE1152 pKa = 3.87WFYY1155 pKa = 11.38QVQGTNNRR1163 pKa = 11.84VSLGDD1168 pKa = 3.62FFEE1171 pKa = 6.18LEE1173 pKa = 4.26LVANTLPGPGIYY1185 pKa = 10.03DD1186 pKa = 3.82IIFVAEE1192 pKa = 4.2DD1193 pKa = 4.54PIVPGCLVEE1202 pKa = 5.13KK1203 pKa = 10.42VVEE1206 pKa = 4.62LIVWPLPAVEE1216 pKa = 5.04AVVLTDD1222 pKa = 5.46ADD1224 pKa = 3.67ACGQPTGSFEE1234 pKa = 4.02ITALTAADD1242 pKa = 4.33EE1243 pKa = 4.78IIVVEE1248 pKa = 4.25TGQRR1252 pKa = 11.84FTNVNAGDD1260 pKa = 3.74VLPIVSNLEE1269 pKa = 3.84PGVYY1273 pKa = 8.31TVRR1276 pKa = 11.84ATNDD1280 pKa = 3.37TGCEE1284 pKa = 3.64FSLPVQIRR1292 pKa = 11.84NLNPPTGIEE1301 pKa = 4.03DD1302 pKa = 4.95FIVQASPEE1310 pKa = 4.1VCGPVGVEE1318 pKa = 3.65NGEE1321 pKa = 4.2LLVVFPAGPQTGTYY1335 pKa = 9.64RR1336 pKa = 11.84AIRR1339 pKa = 11.84VEE1341 pKa = 4.16NGLEE1345 pKa = 3.96TTGAFTNEE1353 pKa = 3.57DD1354 pKa = 3.38SVRR1357 pKa = 11.84ISVEE1361 pKa = 3.3AGTYY1365 pKa = 8.51TIEE1368 pKa = 6.29IIDD1371 pKa = 3.84QFNCSVPDD1379 pKa = 4.07PGQYY1383 pKa = 9.86IVDD1386 pKa = 3.56EE1387 pKa = 4.47RR1388 pKa = 11.84EE1389 pKa = 3.65QAEE1392 pKa = 3.99FSIPNDD1398 pKa = 3.35LSICQEE1404 pKa = 4.02FTFTPVSQQNLVYY1417 pKa = 10.43TLVGPNNQPITPNQDD1432 pKa = 2.24GSYY1435 pKa = 8.62TISVTGQYY1443 pKa = 10.5TMTGTDD1449 pKa = 4.13PNGLLCPAQRR1459 pKa = 11.84QITILNNIFVDD1470 pKa = 4.61FAVSFAQVDD1479 pKa = 4.13CALGQYY1485 pKa = 10.27FDD1487 pKa = 6.09AILNNTNPGNVNFFWRR1503 pKa = 11.84NEE1505 pKa = 3.76AGDD1508 pKa = 3.54IVGRR1512 pKa = 11.84DD1513 pKa = 3.29QRR1515 pKa = 11.84FFPPAPGTYY1524 pKa = 9.85SLEE1527 pKa = 3.98VQPRR1531 pKa = 11.84SGALCPPRR1539 pKa = 11.84IITFTAPPYY1548 pKa = 10.49ALSVDD1553 pKa = 3.56AALEE1557 pKa = 4.16VIPFCVEE1564 pKa = 3.6DD1565 pKa = 3.79AFTVVSLEE1573 pKa = 3.75ADD1575 pKa = 3.22QTFVEE1580 pKa = 4.9RR1581 pKa = 11.84IDD1583 pKa = 3.34WLFVVNNNPVALPQYY1598 pKa = 10.8EE1599 pKa = 4.39NLTSINVSDD1608 pKa = 3.55EE1609 pKa = 4.3GIYY1612 pKa = 10.1QVVLTSVDD1620 pKa = 3.17GCEE1623 pKa = 4.07LARR1626 pKa = 11.84EE1627 pKa = 4.14QIQVIRR1633 pKa = 11.84STIVPPVIDD1642 pKa = 3.91PVYY1645 pKa = 10.11IICAVEE1651 pKa = 4.42GVTATITPGTYY1662 pKa = 8.42STYY1665 pKa = 9.71RR1666 pKa = 11.84WLLNGVEE1673 pKa = 4.3ISTAPEE1679 pKa = 3.92LTPTEE1684 pKa = 4.26AGSYY1688 pKa = 10.21LLTVGDD1694 pKa = 5.43DD1695 pKa = 3.99LGCSYY1700 pKa = 10.55TVSFTVNEE1708 pKa = 4.1DD1709 pKa = 3.21CALRR1713 pKa = 11.84VIFPDD1718 pKa = 4.16AMTPSNPSKK1727 pKa = 10.55RR1728 pKa = 11.84FEE1730 pKa = 4.28VYY1732 pKa = 10.48TNDD1735 pKa = 3.23FVSDD1739 pKa = 3.84LEE1741 pKa = 4.26VFIFNRR1747 pKa = 11.84WGEE1750 pKa = 4.29LIFYY1754 pKa = 7.64CQQSNINGEE1763 pKa = 4.15INVCQWDD1770 pKa = 3.94GTVDD1774 pKa = 3.69GKK1776 pKa = 10.43YY1777 pKa = 10.29VPIGVYY1783 pKa = 9.75PVVVKK1788 pKa = 10.57YY1789 pKa = 10.63RR1790 pKa = 11.84SQRR1793 pKa = 11.84QNVTNSITKK1802 pKa = 10.0SIIVIEE1808 pKa = 4.16
MM1 pKa = 7.44NPTIVFGQGFNNNEE15 pKa = 4.02WVFGDD20 pKa = 4.91CGTGQNNYY28 pKa = 10.36LSFGKK33 pKa = 10.65GGDD36 pKa = 3.82PIVRR40 pKa = 11.84TLPSGVIVGQNNNAVAIDD58 pKa = 4.74PISGNIIFYY67 pKa = 10.7SNGVLVYY74 pKa = 10.37DD75 pKa = 4.71ANNQILEE82 pKa = 4.27GVAPGINGDD91 pKa = 3.56INGQQQVGIGILSYY105 pKa = 10.54EE106 pKa = 4.13PQGDD110 pKa = 3.11RR111 pKa = 11.84LYY113 pKa = 11.06YY114 pKa = 9.84IFYY117 pKa = 10.34VSPAGDD123 pKa = 3.62LQYY126 pKa = 12.09ALIDD130 pKa = 3.72MNAPGQAVGNEE141 pKa = 4.05PPLGEE146 pKa = 5.15IIEE149 pKa = 4.36KK150 pKa = 10.49DD151 pKa = 3.5QVVVGAVNGPILVVKK166 pKa = 9.09TPQSPSYY173 pKa = 10.1LISIEE178 pKa = 3.99GGSIVSRR185 pKa = 11.84RR186 pKa = 11.84IGANPGEE193 pKa = 4.22FTLISNAAVPFTPKK207 pKa = 10.37FMVFDD212 pKa = 3.87EE213 pKa = 4.49ASQRR217 pKa = 11.84VILIPDD223 pKa = 4.32DD224 pKa = 4.12PNQNIVVLGFDD235 pKa = 3.65TASGTFGSQEE245 pKa = 4.47PISQSSNGQAYY256 pKa = 9.81GGASFSPDD264 pKa = 2.54GAFIYY269 pKa = 9.73FSRR272 pKa = 11.84GNQLFRR278 pKa = 11.84IPSNNLTASPEE289 pKa = 4.41AIPLDD294 pKa = 4.03NPVHH298 pKa = 6.93AIYY301 pKa = 10.18DD302 pKa = 3.91VKK304 pKa = 11.03VGPDD308 pKa = 2.85GRR310 pKa = 11.84LYY312 pKa = 10.67YY313 pKa = 9.94IYY315 pKa = 10.49EE316 pKa = 4.15EE317 pKa = 4.33VLGGPQLIGRR327 pKa = 11.84VNNPNEE333 pKa = 4.12PVLDD337 pKa = 4.17DD338 pKa = 4.63LVLAEE343 pKa = 5.25DD344 pKa = 4.7PFNGTDD350 pKa = 3.47FCGRR354 pKa = 11.84IFPKK358 pKa = 10.41FAPNATISPTVDD370 pKa = 4.01FTWEE374 pKa = 3.73PDD376 pKa = 3.63QPCSNNPVQLTSLITPEE393 pKa = 3.69NYY395 pKa = 9.83RR396 pKa = 11.84PVSFEE401 pKa = 3.59WTFNPPLVDD410 pKa = 3.73EE411 pKa = 5.6DD412 pKa = 4.28GQPVDD417 pKa = 3.34IDD419 pKa = 3.57FTQEE423 pKa = 3.47HH424 pKa = 6.7LLIPEE429 pKa = 4.84DD430 pKa = 3.34ATANQSIDD438 pKa = 3.25VTLTVTFADD447 pKa = 4.79GSTQTVNKK455 pKa = 9.24TISLLEE461 pKa = 4.16NDD463 pKa = 4.39LQVQFSSQDD472 pKa = 3.23TTVCEE477 pKa = 4.55GACVDD482 pKa = 3.62IGSMLEE488 pKa = 3.96VQQGGGQGGGQGGGQGNYY506 pKa = 9.45EE507 pKa = 4.24YY508 pKa = 10.0FWSNIRR514 pKa = 11.84EE515 pKa = 4.13WRR517 pKa = 11.84SSKK520 pKa = 10.77DD521 pKa = 3.24NCVDD525 pKa = 3.75LPGLYY530 pKa = 8.72WVLVRR535 pKa = 11.84EE536 pKa = 4.76PGSSCYY542 pKa = 10.37AYY544 pKa = 9.8GSVRR548 pKa = 11.84VRR550 pKa = 11.84IWDD553 pKa = 4.85LLDD556 pKa = 3.11QSNNVWYY563 pKa = 10.11FGDD566 pKa = 4.18GAGLDD571 pKa = 4.3FNPDD575 pKa = 3.63PNNPNAPLPRR585 pKa = 11.84PVGHH589 pKa = 6.26NQNIPAGTTTISDD602 pKa = 3.35GTGQVLFFTDD612 pKa = 5.3GEE614 pKa = 4.97TVWDD618 pKa = 3.94LNGNIMEE625 pKa = 4.92NGDD628 pKa = 4.23SIGGSNLSSQSVIAVPVPTDD648 pKa = 3.07EE649 pKa = 4.55TIFYY653 pKa = 10.89LFTTQLAQDD662 pKa = 4.33GSNQVKK668 pKa = 9.9FSVVDD673 pKa = 3.98IKK675 pKa = 11.45AEE677 pKa = 3.97NQNGVGNVVSKK688 pKa = 11.44DD689 pKa = 3.27NFLFSPSTEE698 pKa = 3.76HH699 pKa = 6.8SAALAAGDD707 pKa = 3.95TTWVLFHH714 pKa = 7.06EE715 pKa = 5.11LGNNTFRR722 pKa = 11.84AYY724 pKa = 9.44PVSIHH729 pKa = 7.26GIGQPVLSSVGSSHH743 pKa = 6.38GFNSGVGSMKK753 pKa = 10.43FSPDD757 pKa = 2.97GTKK760 pKa = 10.44VAVTISDD767 pKa = 4.35GACSRR772 pKa = 11.84LEE774 pKa = 4.37IFDD777 pKa = 4.46FDD779 pKa = 4.15QNTGEE784 pKa = 4.09MTEE787 pKa = 4.12YY788 pKa = 11.42ALLDD792 pKa = 3.97LGCNNDD798 pKa = 3.11QVYY801 pKa = 10.72GLEE804 pKa = 3.99FSNDD808 pKa = 2.81GSRR811 pKa = 11.84VYY813 pKa = 10.81VSYY816 pKa = 10.92RR817 pKa = 11.84GGNGKK822 pKa = 8.93IDD824 pKa = 3.76EE825 pKa = 4.75FLIQAPEE832 pKa = 3.86QTGNNPIPCATCFSSATTRR851 pKa = 11.84PQRR854 pKa = 11.84EE855 pKa = 3.86ACILNSPVRR864 pKa = 11.84NTLTTSGPFGALQIGPDD881 pKa = 3.55GQIYY885 pKa = 8.59VARR888 pKa = 11.84PGQNVLGTINAGQDD902 pKa = 4.0CNRR905 pKa = 11.84SAYY908 pKa = 10.04NEE910 pKa = 4.32NGTSTLLGTSNLGLPSFVQQSGSSIPEE937 pKa = 3.75PALAGTPFLCIDD949 pKa = 4.06PQNGALGEE957 pKa = 4.19FEE959 pKa = 5.44GGGEE963 pKa = 4.17PDD965 pKa = 2.93IDD967 pKa = 3.99SYY969 pKa = 11.62FWTIVHH975 pKa = 6.9EE976 pKa = 5.02DD977 pKa = 3.29GTSILNNYY985 pKa = 8.14GGPGEE990 pKa = 4.03NFQVLEE996 pKa = 4.2QPFTRR1001 pKa = 11.84PGTYY1005 pKa = 8.87TVSLRR1010 pKa = 11.84VDD1012 pKa = 3.06RR1013 pKa = 11.84CGDD1016 pKa = 3.37PDD1018 pKa = 4.01FYY1020 pKa = 11.05EE1021 pKa = 4.66ASLEE1025 pKa = 4.27VEE1027 pKa = 4.88VIAPPTLTLPGEE1039 pKa = 4.3ATLCSGNPVSLTAIDD1054 pKa = 5.26GYY1056 pKa = 11.51DD1057 pKa = 3.76PADD1060 pKa = 3.52GLYY1063 pKa = 10.83DD1064 pKa = 3.84FEE1066 pKa = 4.45WRR1068 pKa = 11.84NAQGTLFGDD1077 pKa = 4.11SNSNTIFVNEE1087 pKa = 3.94EE1088 pKa = 4.07SIFTVTVSHH1097 pKa = 7.0RR1098 pKa = 11.84RR1099 pKa = 11.84PDD1101 pKa = 4.52DD1102 pKa = 3.37IPAEE1106 pKa = 4.11FFNPCPATASVFVGPPFDD1124 pKa = 5.08FDD1126 pKa = 3.71LTQTADD1132 pKa = 3.26QVCFDD1137 pKa = 3.76EE1138 pKa = 4.72SLVVFAPNTPIVGEE1152 pKa = 3.87WFYY1155 pKa = 11.38QVQGTNNRR1163 pKa = 11.84VSLGDD1168 pKa = 3.62FFEE1171 pKa = 6.18LEE1173 pKa = 4.26LVANTLPGPGIYY1185 pKa = 10.03DD1186 pKa = 3.82IIFVAEE1192 pKa = 4.2DD1193 pKa = 4.54PIVPGCLVEE1202 pKa = 5.13KK1203 pKa = 10.42VVEE1206 pKa = 4.62LIVWPLPAVEE1216 pKa = 5.04AVVLTDD1222 pKa = 5.46ADD1224 pKa = 3.67ACGQPTGSFEE1234 pKa = 4.02ITALTAADD1242 pKa = 4.33EE1243 pKa = 4.78IIVVEE1248 pKa = 4.25TGQRR1252 pKa = 11.84FTNVNAGDD1260 pKa = 3.74VLPIVSNLEE1269 pKa = 3.84PGVYY1273 pKa = 8.31TVRR1276 pKa = 11.84ATNDD1280 pKa = 3.37TGCEE1284 pKa = 3.64FSLPVQIRR1292 pKa = 11.84NLNPPTGIEE1301 pKa = 4.03DD1302 pKa = 4.95FIVQASPEE1310 pKa = 4.1VCGPVGVEE1318 pKa = 3.65NGEE1321 pKa = 4.2LLVVFPAGPQTGTYY1335 pKa = 9.64RR1336 pKa = 11.84AIRR1339 pKa = 11.84VEE1341 pKa = 4.16NGLEE1345 pKa = 3.96TTGAFTNEE1353 pKa = 3.57DD1354 pKa = 3.38SVRR1357 pKa = 11.84ISVEE1361 pKa = 3.3AGTYY1365 pKa = 8.51TIEE1368 pKa = 6.29IIDD1371 pKa = 3.84QFNCSVPDD1379 pKa = 4.07PGQYY1383 pKa = 9.86IVDD1386 pKa = 3.56EE1387 pKa = 4.47RR1388 pKa = 11.84EE1389 pKa = 3.65QAEE1392 pKa = 3.99FSIPNDD1398 pKa = 3.35LSICQEE1404 pKa = 4.02FTFTPVSQQNLVYY1417 pKa = 10.43TLVGPNNQPITPNQDD1432 pKa = 2.24GSYY1435 pKa = 8.62TISVTGQYY1443 pKa = 10.5TMTGTDD1449 pKa = 4.13PNGLLCPAQRR1459 pKa = 11.84QITILNNIFVDD1470 pKa = 4.61FAVSFAQVDD1479 pKa = 4.13CALGQYY1485 pKa = 10.27FDD1487 pKa = 6.09AILNNTNPGNVNFFWRR1503 pKa = 11.84NEE1505 pKa = 3.76AGDD1508 pKa = 3.54IVGRR1512 pKa = 11.84DD1513 pKa = 3.29QRR1515 pKa = 11.84FFPPAPGTYY1524 pKa = 9.85SLEE1527 pKa = 3.98VQPRR1531 pKa = 11.84SGALCPPRR1539 pKa = 11.84IITFTAPPYY1548 pKa = 10.49ALSVDD1553 pKa = 3.56AALEE1557 pKa = 4.16VIPFCVEE1564 pKa = 3.6DD1565 pKa = 3.79AFTVVSLEE1573 pKa = 3.75ADD1575 pKa = 3.22QTFVEE1580 pKa = 4.9RR1581 pKa = 11.84IDD1583 pKa = 3.34WLFVVNNNPVALPQYY1598 pKa = 10.8EE1599 pKa = 4.39NLTSINVSDD1608 pKa = 3.55EE1609 pKa = 4.3GIYY1612 pKa = 10.1QVVLTSVDD1620 pKa = 3.17GCEE1623 pKa = 4.07LARR1626 pKa = 11.84EE1627 pKa = 4.14QIQVIRR1633 pKa = 11.84STIVPPVIDD1642 pKa = 3.91PVYY1645 pKa = 10.11IICAVEE1651 pKa = 4.42GVTATITPGTYY1662 pKa = 8.42STYY1665 pKa = 9.71RR1666 pKa = 11.84WLLNGVEE1673 pKa = 4.3ISTAPEE1679 pKa = 3.92LTPTEE1684 pKa = 4.26AGSYY1688 pKa = 10.21LLTVGDD1694 pKa = 5.43DD1695 pKa = 3.99LGCSYY1700 pKa = 10.55TVSFTVNEE1708 pKa = 4.1DD1709 pKa = 3.21CALRR1713 pKa = 11.84VIFPDD1718 pKa = 4.16AMTPSNPSKK1727 pKa = 10.55RR1728 pKa = 11.84FEE1730 pKa = 4.28VYY1732 pKa = 10.48TNDD1735 pKa = 3.23FVSDD1739 pKa = 3.84LEE1741 pKa = 4.26VFIFNRR1747 pKa = 11.84WGEE1750 pKa = 4.29LIFYY1754 pKa = 7.64CQQSNINGEE1763 pKa = 4.15INVCQWDD1770 pKa = 3.94GTVDD1774 pKa = 3.69GKK1776 pKa = 10.43YY1777 pKa = 10.29VPIGVYY1783 pKa = 9.75PVVVKK1788 pKa = 10.57YY1789 pKa = 10.63RR1790 pKa = 11.84SQRR1793 pKa = 11.84QNVTNSITKK1802 pKa = 10.0SIIVIEE1808 pKa = 4.16
Molecular weight: 195.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7XA83|M7XA83_9BACT GCN5-related N-acetyltransferase OS=Mariniradius saccharolyticus AK6 OX=1239962 GN=C943_03544 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1435942 |
37 |
4749 |
309.9 |
34.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.076 ± 0.035 | 0.679 ± 0.011 |
5.3 ± 0.026 | 6.607 ± 0.034 |
5.514 ± 0.031 | 7.302 ± 0.034 |
1.896 ± 0.02 | 6.928 ± 0.028 |
6.587 ± 0.038 | 9.722 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.508 ± 0.018 | 4.811 ± 0.028 |
4.173 ± 0.022 | 3.735 ± 0.018 |
4.436 ± 0.024 | 6.265 ± 0.026 |
5.042 ± 0.031 | 6.478 ± 0.031 |
1.347 ± 0.013 | 3.592 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
