
Cryptobacterium curtum (strain ATCC 700683 / DSM 15641 / CCUG 43107 / 12-3)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Cryptobacterium; Cryptobacterium curtum
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
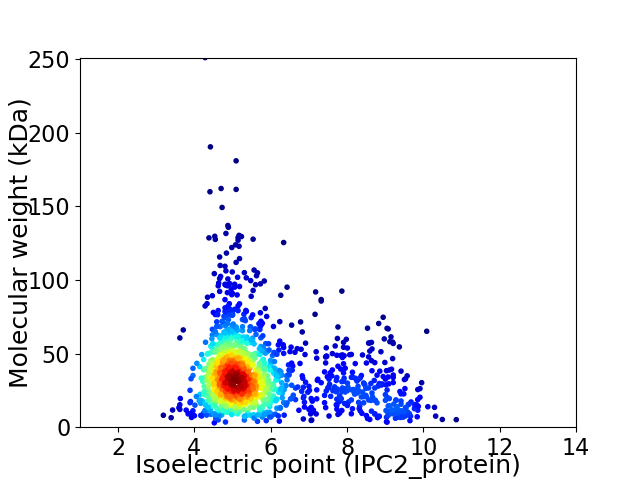
Virtual 2D-PAGE plot for 1355 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
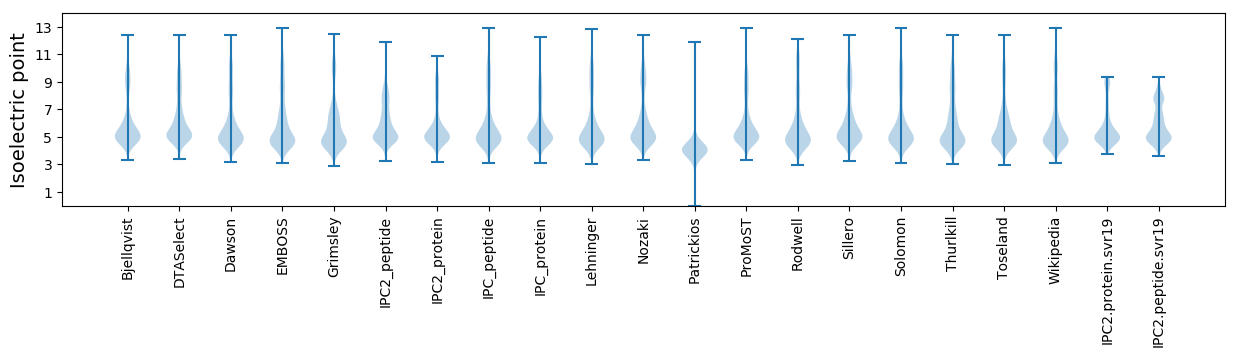
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7MMP9|C7MMP9_CRYCD Uncharacterized protein OS=Cryptobacterium curtum (strain ATCC 700683 / DSM 15641 / CCUG 43107 / 12-3) OX=469378 GN=Ccur_04670 PE=4 SV=1
MM1 pKa = 7.85KK2 pKa = 9.95DD3 pKa = 3.45ARR5 pKa = 11.84WRR7 pKa = 11.84WIATAACCVLIVVSAWTLGSYY28 pKa = 10.71FLGSSGASEE37 pKa = 5.09DD38 pKa = 4.2LFTTVAPAEE47 pKa = 4.28SDD49 pKa = 3.04ASSGVTTTGDD59 pKa = 4.7LIPKK63 pKa = 9.86QEE65 pKa = 5.52DD66 pKa = 3.38DD67 pKa = 4.29TADD70 pKa = 3.49QSDD73 pKa = 3.93GAAGEE78 pKa = 4.55DD79 pKa = 3.9TGSSDD84 pKa = 5.59DD85 pKa = 3.91PTPPLAAADD94 pKa = 3.72SAYY97 pKa = 9.51ATQSGDD103 pKa = 3.6PQSSDD108 pKa = 2.95SSSRR112 pKa = 11.84SSDD115 pKa = 3.05GSSVAGGSSASSATTSGSSSSSSSSGDD142 pKa = 3.28QLTHH146 pKa = 5.36IQVNITVDD154 pKa = 3.52GTAAGSNISSALLSIPAGSTVYY176 pKa = 10.58DD177 pKa = 3.63ALKK180 pKa = 9.81ATGASVNARR189 pKa = 11.84STVYY193 pKa = 10.87GMYY196 pKa = 10.24VAGINGLAEE205 pKa = 4.29KK206 pKa = 10.26EE207 pKa = 4.08YY208 pKa = 10.5GAQSGWIYY216 pKa = 10.57YY217 pKa = 10.55VNGQFADD224 pKa = 4.29RR225 pKa = 11.84ACDD228 pKa = 2.99RR229 pKa = 11.84WRR231 pKa = 11.84LSEE234 pKa = 3.86GDD236 pKa = 3.74YY237 pKa = 11.14VKK239 pKa = 10.11WVYY242 pKa = 11.04VSDD245 pKa = 3.7NN246 pKa = 3.35
MM1 pKa = 7.85KK2 pKa = 9.95DD3 pKa = 3.45ARR5 pKa = 11.84WRR7 pKa = 11.84WIATAACCVLIVVSAWTLGSYY28 pKa = 10.71FLGSSGASEE37 pKa = 5.09DD38 pKa = 4.2LFTTVAPAEE47 pKa = 4.28SDD49 pKa = 3.04ASSGVTTTGDD59 pKa = 4.7LIPKK63 pKa = 9.86QEE65 pKa = 5.52DD66 pKa = 3.38DD67 pKa = 4.29TADD70 pKa = 3.49QSDD73 pKa = 3.93GAAGEE78 pKa = 4.55DD79 pKa = 3.9TGSSDD84 pKa = 5.59DD85 pKa = 3.91PTPPLAAADD94 pKa = 3.72SAYY97 pKa = 9.51ATQSGDD103 pKa = 3.6PQSSDD108 pKa = 2.95SSSRR112 pKa = 11.84SSDD115 pKa = 3.05GSSVAGGSSASSATTSGSSSSSSSSGDD142 pKa = 3.28QLTHH146 pKa = 5.36IQVNITVDD154 pKa = 3.52GTAAGSNISSALLSIPAGSTVYY176 pKa = 10.58DD177 pKa = 3.63ALKK180 pKa = 9.81ATGASVNARR189 pKa = 11.84STVYY193 pKa = 10.87GMYY196 pKa = 10.24VAGINGLAEE205 pKa = 4.29KK206 pKa = 10.26EE207 pKa = 4.08YY208 pKa = 10.5GAQSGWIYY216 pKa = 10.57YY217 pKa = 10.55VNGQFADD224 pKa = 4.29RR225 pKa = 11.84ACDD228 pKa = 2.99RR229 pKa = 11.84WRR231 pKa = 11.84LSEE234 pKa = 3.86GDD236 pKa = 3.74YY237 pKa = 11.14VKK239 pKa = 10.11WVYY242 pKa = 11.04VSDD245 pKa = 3.7NN246 pKa = 3.35
Molecular weight: 25.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7MMP7|C7MMP7_CRYCD ABC-type cobalt transport system ATPase component OS=Cryptobacterium curtum (strain ATCC 700683 / DSM 15641 / CCUG 43107 / 12-3) OX=469378 GN=Ccur_04650 PE=4 SV=1
MM1 pKa = 7.6ARR3 pKa = 11.84RR4 pKa = 11.84TSEE7 pKa = 3.76HH8 pKa = 6.15EE9 pKa = 3.64ARR11 pKa = 11.84ILDD14 pKa = 3.73FEE16 pKa = 4.41EE17 pKa = 3.89ARR19 pKa = 11.84RR20 pKa = 11.84PARR23 pKa = 11.84EE24 pKa = 3.49QSTRR28 pKa = 11.84AARR31 pKa = 11.84PSTRR35 pKa = 11.84RR36 pKa = 11.84ATRR39 pKa = 11.84TASSRR44 pKa = 11.84SSARR48 pKa = 11.84GEE50 pKa = 4.33SSSQPARR57 pKa = 11.84RR58 pKa = 11.84STGAASSSQAGSVRR72 pKa = 11.84RR73 pKa = 11.84SGRR76 pKa = 11.84RR77 pKa = 11.84IQSSARR83 pKa = 11.84EE84 pKa = 3.84EE85 pKa = 4.45SFCRR89 pKa = 11.84SASTASSRR97 pKa = 11.84HH98 pKa = 3.9STRR101 pKa = 11.84PHH103 pKa = 5.21VSARR107 pKa = 11.84QQQSGRR113 pKa = 11.84ITSFFKK119 pKa = 10.83GLQKK123 pKa = 10.52KK124 pKa = 10.1RR125 pKa = 11.84LDD127 pKa = 3.38KK128 pKa = 11.23RR129 pKa = 11.84FDD131 pKa = 3.42RR132 pKa = 11.84SVRR135 pKa = 11.84TGDD138 pKa = 3.59DD139 pKa = 3.45TTHH142 pKa = 6.65EE143 pKa = 4.62SSNSARR149 pKa = 11.84PKK151 pKa = 8.79AALYY155 pKa = 6.58STHH158 pKa = 6.5MGRR161 pKa = 11.84VHH163 pKa = 6.9RR164 pKa = 11.84KK165 pKa = 7.55SSRR168 pKa = 11.84MQNTAARR175 pKa = 11.84TTSRR179 pKa = 11.84AGSASIATSSRR190 pKa = 11.84QRR192 pKa = 11.84WVVPVCVVAFLVLCGAFLYY211 pKa = 10.87APAKK215 pKa = 10.2DD216 pKa = 4.0YY217 pKa = 11.54YY218 pKa = 10.18IASRR222 pKa = 11.84DD223 pKa = 3.54AAKK226 pKa = 8.92TQAEE230 pKa = 4.5YY231 pKa = 11.39NRR233 pKa = 11.84VSQEE237 pKa = 3.62NSQLQSDD244 pKa = 4.61VEE246 pKa = 4.38SLSTDD251 pKa = 2.91EE252 pKa = 5.39GIEE255 pKa = 3.78DD256 pKa = 4.36RR257 pKa = 11.84ARR259 pKa = 11.84EE260 pKa = 3.85DD261 pKa = 3.56YY262 pKa = 11.07GWVKK266 pKa = 10.59EE267 pKa = 4.29GEE269 pKa = 4.23NSVSVSGLSSDD280 pKa = 4.65RR281 pKa = 11.84DD282 pKa = 3.46QSTNASSKK290 pKa = 10.89ADD292 pKa = 3.25AASEE296 pKa = 4.39VKK298 pKa = 10.69APTTWYY304 pKa = 10.75SGILDD309 pKa = 3.77AFFGYY314 pKa = 10.26KK315 pKa = 10.28DD316 pKa = 3.36SS317 pKa = 3.98
MM1 pKa = 7.6ARR3 pKa = 11.84RR4 pKa = 11.84TSEE7 pKa = 3.76HH8 pKa = 6.15EE9 pKa = 3.64ARR11 pKa = 11.84ILDD14 pKa = 3.73FEE16 pKa = 4.41EE17 pKa = 3.89ARR19 pKa = 11.84RR20 pKa = 11.84PARR23 pKa = 11.84EE24 pKa = 3.49QSTRR28 pKa = 11.84AARR31 pKa = 11.84PSTRR35 pKa = 11.84RR36 pKa = 11.84ATRR39 pKa = 11.84TASSRR44 pKa = 11.84SSARR48 pKa = 11.84GEE50 pKa = 4.33SSSQPARR57 pKa = 11.84RR58 pKa = 11.84STGAASSSQAGSVRR72 pKa = 11.84RR73 pKa = 11.84SGRR76 pKa = 11.84RR77 pKa = 11.84IQSSARR83 pKa = 11.84EE84 pKa = 3.84EE85 pKa = 4.45SFCRR89 pKa = 11.84SASTASSRR97 pKa = 11.84HH98 pKa = 3.9STRR101 pKa = 11.84PHH103 pKa = 5.21VSARR107 pKa = 11.84QQQSGRR113 pKa = 11.84ITSFFKK119 pKa = 10.83GLQKK123 pKa = 10.52KK124 pKa = 10.1RR125 pKa = 11.84LDD127 pKa = 3.38KK128 pKa = 11.23RR129 pKa = 11.84FDD131 pKa = 3.42RR132 pKa = 11.84SVRR135 pKa = 11.84TGDD138 pKa = 3.59DD139 pKa = 3.45TTHH142 pKa = 6.65EE143 pKa = 4.62SSNSARR149 pKa = 11.84PKK151 pKa = 8.79AALYY155 pKa = 6.58STHH158 pKa = 6.5MGRR161 pKa = 11.84VHH163 pKa = 6.9RR164 pKa = 11.84KK165 pKa = 7.55SSRR168 pKa = 11.84MQNTAARR175 pKa = 11.84TTSRR179 pKa = 11.84AGSASIATSSRR190 pKa = 11.84QRR192 pKa = 11.84WVVPVCVVAFLVLCGAFLYY211 pKa = 10.87APAKK215 pKa = 10.2DD216 pKa = 4.0YY217 pKa = 11.54YY218 pKa = 10.18IASRR222 pKa = 11.84DD223 pKa = 3.54AAKK226 pKa = 8.92TQAEE230 pKa = 4.5YY231 pKa = 11.39NRR233 pKa = 11.84VSQEE237 pKa = 3.62NSQLQSDD244 pKa = 4.61VEE246 pKa = 4.38SLSTDD251 pKa = 2.91EE252 pKa = 5.39GIEE255 pKa = 3.78DD256 pKa = 4.36RR257 pKa = 11.84ARR259 pKa = 11.84EE260 pKa = 3.85DD261 pKa = 3.56YY262 pKa = 11.07GWVKK266 pKa = 10.59EE267 pKa = 4.29GEE269 pKa = 4.23NSVSVSGLSSDD280 pKa = 4.65RR281 pKa = 11.84DD282 pKa = 3.46QSTNASSKK290 pKa = 10.89ADD292 pKa = 3.25AASEE296 pKa = 4.39VKK298 pKa = 10.69APTTWYY304 pKa = 10.75SGILDD309 pKa = 3.77AFFGYY314 pKa = 10.26KK315 pKa = 10.28DD316 pKa = 3.36SS317 pKa = 3.98
Molecular weight: 34.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
470126 |
26 |
2400 |
347.0 |
37.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.275 ± 0.078 | 1.503 ± 0.032 |
6.109 ± 0.061 | 6.019 ± 0.063 |
3.702 ± 0.045 | 7.632 ± 0.056 |
2.106 ± 0.028 | 5.81 ± 0.053 |
3.746 ± 0.052 | 8.976 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.527 ± 0.033 | 3.122 ± 0.044 |
4.106 ± 0.039 | 3.538 ± 0.042 |
5.665 ± 0.061 | 6.689 ± 0.069 |
5.92 ± 0.07 | 7.754 ± 0.054 |
1.042 ± 0.024 | 2.757 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
