
Microbacterium ketosireducens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia;
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
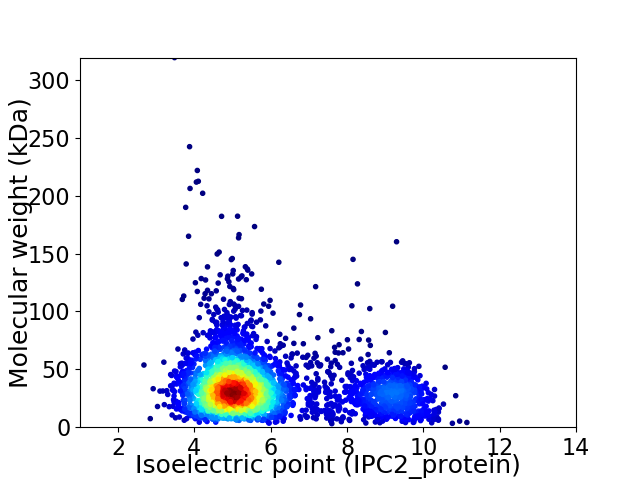
Virtual 2D-PAGE plot for 3448 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M2GVJ6|A0A0M2GVJ6_9MICO Major Facilitator Superfamily protein OS=Microbacterium ketosireducens OX=92835 GN=RS81_03299 PE=4 SV=1
MM1 pKa = 7.33FRR3 pKa = 11.84RR4 pKa = 11.84SLIAVTAAATALVLASCSAGGNPEE28 pKa = 3.97PTDD31 pKa = 3.88DD32 pKa = 3.4EE33 pKa = 4.4AARR36 pKa = 11.84GDD38 pKa = 4.04RR39 pKa = 11.84LTLISISSPTNYY51 pKa = 10.4DD52 pKa = 3.33LGVGAEE58 pKa = 4.07AGTRR62 pKa = 11.84SQFFEE67 pKa = 4.36AVFDD71 pKa = 3.86TLLRR75 pKa = 11.84ADD77 pKa = 3.76SAGEE81 pKa = 3.64ISPRR85 pKa = 11.84LATDD89 pKa = 3.62WSYY92 pKa = 12.27DD93 pKa = 3.54DD94 pKa = 4.34TLTEE98 pKa = 3.85LTLTLRR104 pKa = 11.84EE105 pKa = 4.48DD106 pKa = 3.58VTFSDD111 pKa = 4.29GTALDD116 pKa = 3.58AAAVVASLEE125 pKa = 4.15HH126 pKa = 5.99TRR128 pKa = 11.84DD129 pKa = 3.44GSSQAAGAAVGQTYY143 pKa = 9.98EE144 pKa = 4.35AVDD147 pKa = 4.26DD148 pKa = 4.02YY149 pKa = 10.72TVKK152 pKa = 9.4ITLPAPNPSYY162 pKa = 10.68LGSLAVTSGLIAAPSSFDD180 pKa = 3.94NDD182 pKa = 4.13DD183 pKa = 3.22VDD185 pKa = 4.31TNPVGSGPFILDD197 pKa = 3.38TEE199 pKa = 4.59STVIGTTYY207 pKa = 10.97NYY209 pKa = 7.65TANPDD214 pKa = 3.22YY215 pKa = 10.27WDD217 pKa = 4.47PEE219 pKa = 4.52SIRR222 pKa = 11.84YY223 pKa = 8.34DD224 pKa = 3.24HH225 pKa = 6.47LTINTIADD233 pKa = 3.39PTATLNAILAGEE245 pKa = 4.4TNGAVADD252 pKa = 4.01NNALEE257 pKa = 4.67EE258 pKa = 4.23IEE260 pKa = 4.17GAGWDD265 pKa = 3.78FTEE268 pKa = 6.35SEE270 pKa = 4.59GAWEE274 pKa = 4.55GLMLYY279 pKa = 10.59DD280 pKa = 4.92RR281 pKa = 11.84DD282 pKa = 3.81GAMAPALADD291 pKa = 3.21VRR293 pKa = 11.84VRR295 pKa = 11.84QAINMAFDD303 pKa = 3.82RR304 pKa = 11.84EE305 pKa = 4.37SLLEE309 pKa = 4.03AVQGGHH315 pKa = 5.38GTVTTQIFMPNTSTYY330 pKa = 11.0DD331 pKa = 3.49PEE333 pKa = 4.74LDD335 pKa = 3.86KK336 pKa = 10.88MYY338 pKa = 10.61AYY340 pKa = 10.4DD341 pKa = 3.8PEE343 pKa = 4.65GAKK346 pKa = 10.33EE347 pKa = 4.0LLTEE351 pKa = 4.66AGYY354 pKa = 9.92PDD356 pKa = 4.96GFTMTMPLIAAFQTSLDD373 pKa = 3.84LLKK376 pKa = 10.7QQLADD381 pKa = 3.32VGITVEE387 pKa = 4.55YY388 pKa = 10.35VDD390 pKa = 4.86PGQGQYY396 pKa = 9.68IASILTPKK404 pKa = 10.4HH405 pKa = 4.87PAAWMNMAGIGDD417 pKa = 4.07WDD419 pKa = 4.59AIGYY423 pKa = 7.07MVSPAAVFNPFRR435 pKa = 11.84TQQDD439 pKa = 3.47QVDD442 pKa = 3.54DD443 pKa = 3.93WLYY446 pKa = 10.59EE447 pKa = 3.97IQNATDD453 pKa = 3.37EE454 pKa = 4.97ADD456 pKa = 4.46ASMTDD461 pKa = 3.37LKK463 pKa = 11.24AYY465 pKa = 10.08LVEE468 pKa = 4.41EE469 pKa = 4.01AWFAPFYY476 pKa = 10.39RR477 pKa = 11.84VNGNYY482 pKa = 7.36TTDD485 pKa = 3.39AEE487 pKa = 4.27TDD489 pKa = 3.4YY490 pKa = 11.57DD491 pKa = 3.76ASTWVSGAYY500 pKa = 9.46PSIFGFSPASS510 pKa = 3.29
MM1 pKa = 7.33FRR3 pKa = 11.84RR4 pKa = 11.84SLIAVTAAATALVLASCSAGGNPEE28 pKa = 3.97PTDD31 pKa = 3.88DD32 pKa = 3.4EE33 pKa = 4.4AARR36 pKa = 11.84GDD38 pKa = 4.04RR39 pKa = 11.84LTLISISSPTNYY51 pKa = 10.4DD52 pKa = 3.33LGVGAEE58 pKa = 4.07AGTRR62 pKa = 11.84SQFFEE67 pKa = 4.36AVFDD71 pKa = 3.86TLLRR75 pKa = 11.84ADD77 pKa = 3.76SAGEE81 pKa = 3.64ISPRR85 pKa = 11.84LATDD89 pKa = 3.62WSYY92 pKa = 12.27DD93 pKa = 3.54DD94 pKa = 4.34TLTEE98 pKa = 3.85LTLTLRR104 pKa = 11.84EE105 pKa = 4.48DD106 pKa = 3.58VTFSDD111 pKa = 4.29GTALDD116 pKa = 3.58AAAVVASLEE125 pKa = 4.15HH126 pKa = 5.99TRR128 pKa = 11.84DD129 pKa = 3.44GSSQAAGAAVGQTYY143 pKa = 9.98EE144 pKa = 4.35AVDD147 pKa = 4.26DD148 pKa = 4.02YY149 pKa = 10.72TVKK152 pKa = 9.4ITLPAPNPSYY162 pKa = 10.68LGSLAVTSGLIAAPSSFDD180 pKa = 3.94NDD182 pKa = 4.13DD183 pKa = 3.22VDD185 pKa = 4.31TNPVGSGPFILDD197 pKa = 3.38TEE199 pKa = 4.59STVIGTTYY207 pKa = 10.97NYY209 pKa = 7.65TANPDD214 pKa = 3.22YY215 pKa = 10.27WDD217 pKa = 4.47PEE219 pKa = 4.52SIRR222 pKa = 11.84YY223 pKa = 8.34DD224 pKa = 3.24HH225 pKa = 6.47LTINTIADD233 pKa = 3.39PTATLNAILAGEE245 pKa = 4.4TNGAVADD252 pKa = 4.01NNALEE257 pKa = 4.67EE258 pKa = 4.23IEE260 pKa = 4.17GAGWDD265 pKa = 3.78FTEE268 pKa = 6.35SEE270 pKa = 4.59GAWEE274 pKa = 4.55GLMLYY279 pKa = 10.59DD280 pKa = 4.92RR281 pKa = 11.84DD282 pKa = 3.81GAMAPALADD291 pKa = 3.21VRR293 pKa = 11.84VRR295 pKa = 11.84QAINMAFDD303 pKa = 3.82RR304 pKa = 11.84EE305 pKa = 4.37SLLEE309 pKa = 4.03AVQGGHH315 pKa = 5.38GTVTTQIFMPNTSTYY330 pKa = 11.0DD331 pKa = 3.49PEE333 pKa = 4.74LDD335 pKa = 3.86KK336 pKa = 10.88MYY338 pKa = 10.61AYY340 pKa = 10.4DD341 pKa = 3.8PEE343 pKa = 4.65GAKK346 pKa = 10.33EE347 pKa = 4.0LLTEE351 pKa = 4.66AGYY354 pKa = 9.92PDD356 pKa = 4.96GFTMTMPLIAAFQTSLDD373 pKa = 3.84LLKK376 pKa = 10.7QQLADD381 pKa = 3.32VGITVEE387 pKa = 4.55YY388 pKa = 10.35VDD390 pKa = 4.86PGQGQYY396 pKa = 9.68IASILTPKK404 pKa = 10.4HH405 pKa = 4.87PAAWMNMAGIGDD417 pKa = 4.07WDD419 pKa = 4.59AIGYY423 pKa = 7.07MVSPAAVFNPFRR435 pKa = 11.84TQQDD439 pKa = 3.47QVDD442 pKa = 3.54DD443 pKa = 3.93WLYY446 pKa = 10.59EE447 pKa = 3.97IQNATDD453 pKa = 3.37EE454 pKa = 4.97ADD456 pKa = 4.46ASMTDD461 pKa = 3.37LKK463 pKa = 11.24AYY465 pKa = 10.08LVEE468 pKa = 4.41EE469 pKa = 4.01AWFAPFYY476 pKa = 10.39RR477 pKa = 11.84VNGNYY482 pKa = 7.36TTDD485 pKa = 3.39AEE487 pKa = 4.27TDD489 pKa = 3.4YY490 pKa = 11.57DD491 pKa = 3.76ASTWVSGAYY500 pKa = 9.46PSIFGFSPASS510 pKa = 3.29
Molecular weight: 54.71 kDa
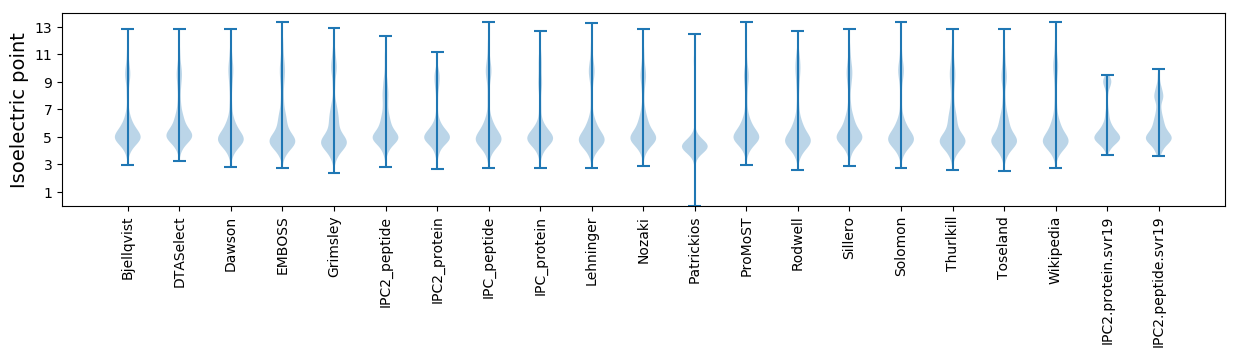
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M2H233|A0A0M2H233_9MICO Putative inactive lipase OS=Microbacterium ketosireducens OX=92835 GN=RS81_03385 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1191655 |
29 |
3088 |
345.6 |
36.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.201 ± 0.063 | 0.467 ± 0.01 |
6.586 ± 0.038 | 5.493 ± 0.041 |
3.145 ± 0.027 | 8.985 ± 0.032 |
1.966 ± 0.022 | 4.539 ± 0.034 |
1.711 ± 0.025 | 9.831 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.759 ± 0.019 | 1.789 ± 0.024 |
5.43 ± 0.028 | 2.578 ± 0.018 |
7.219 ± 0.05 | 5.426 ± 0.028 |
6.124 ± 0.042 | 9.156 ± 0.042 |
1.603 ± 0.019 | 1.992 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
