
Pseudogymnoascus verrucosus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
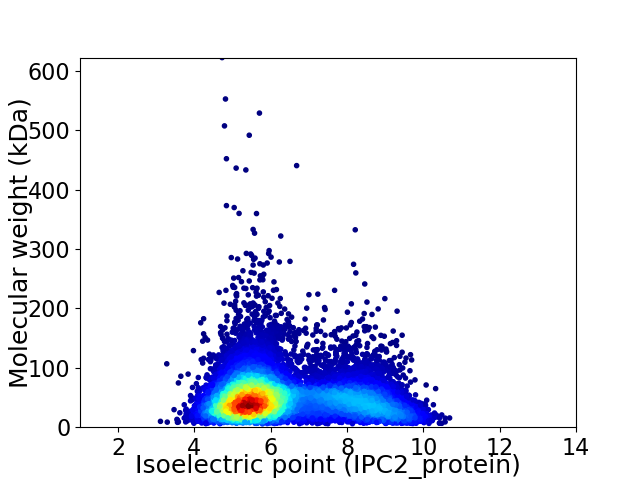
Virtual 2D-PAGE plot for 10683 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B8GDJ5|A0A1B8GDJ5_9PEZI Zn(2)-C6 fungal-type domain-containing protein OS=Pseudogymnoascus verrucosus OX=342668 GN=VE01_08199 PE=4 SV=1
MM1 pKa = 7.54SGYY4 pKa = 11.32GNDD7 pKa = 3.77NNDD10 pKa = 3.5SYY12 pKa = 11.89GSSNTRR18 pKa = 11.84GGDD21 pKa = 3.47DD22 pKa = 4.52SYY24 pKa = 12.11GSSNNNSSNNDD35 pKa = 3.08SYY37 pKa = 11.9GSSNTRR43 pKa = 11.84SSDD46 pKa = 3.38NNDD49 pKa = 3.1SYY51 pKa = 11.93GSSNTRR57 pKa = 11.84SSDD60 pKa = 3.38NNDD63 pKa = 3.12SYY65 pKa = 12.05GSSNKK70 pKa = 9.68NSNDD74 pKa = 3.2DD75 pKa = 3.71DD76 pKa = 5.31SYY78 pKa = 11.7GSSNKK83 pKa = 9.9SSGNTDD89 pKa = 2.93SYY91 pKa = 11.57GSSGNSRR98 pKa = 11.84GDD100 pKa = 3.37NNDD103 pKa = 3.19SYY105 pKa = 11.95GSSNKK110 pKa = 9.82SSSNDD115 pKa = 2.74NDD117 pKa = 4.17TYY119 pKa = 11.56GSSNKK124 pKa = 9.56NSSDD128 pKa = 3.53TYY130 pKa = 11.12GSSNKK135 pKa = 9.55NSSDD139 pKa = 3.67SYY141 pKa = 10.85GSSGNTDD148 pKa = 2.99SYY150 pKa = 11.83GSSNKK155 pKa = 9.68NSNDD159 pKa = 3.04NDD161 pKa = 3.66DD162 pKa = 4.86SYY164 pKa = 12.21GSSNKK169 pKa = 9.79SSSNTDD175 pKa = 2.7SYY177 pKa = 11.94GSSNKK182 pKa = 9.58NSSDD186 pKa = 3.67SYY188 pKa = 10.85GSSGNTDD195 pKa = 2.99SYY197 pKa = 11.83GSSNKK202 pKa = 9.68NSNDD206 pKa = 3.2DD207 pKa = 3.71DD208 pKa = 5.25SYY210 pKa = 11.7GSSNTRR216 pKa = 11.84SSGNNNNSSDD226 pKa = 4.12SYY228 pKa = 11.61GSSNNNNNDD237 pKa = 3.11SYY239 pKa = 12.1GSGNNNSGSSTVDD252 pKa = 3.16KK253 pKa = 11.16LVDD256 pKa = 3.35TATGFLGRR264 pKa = 11.84SGGNNNNNDD273 pKa = 3.15SNYY276 pKa = 10.97
MM1 pKa = 7.54SGYY4 pKa = 11.32GNDD7 pKa = 3.77NNDD10 pKa = 3.5SYY12 pKa = 11.89GSSNTRR18 pKa = 11.84GGDD21 pKa = 3.47DD22 pKa = 4.52SYY24 pKa = 12.11GSSNNNSSNNDD35 pKa = 3.08SYY37 pKa = 11.9GSSNTRR43 pKa = 11.84SSDD46 pKa = 3.38NNDD49 pKa = 3.1SYY51 pKa = 11.93GSSNTRR57 pKa = 11.84SSDD60 pKa = 3.38NNDD63 pKa = 3.12SYY65 pKa = 12.05GSSNKK70 pKa = 9.68NSNDD74 pKa = 3.2DD75 pKa = 3.71DD76 pKa = 5.31SYY78 pKa = 11.7GSSNKK83 pKa = 9.9SSGNTDD89 pKa = 2.93SYY91 pKa = 11.57GSSGNSRR98 pKa = 11.84GDD100 pKa = 3.37NNDD103 pKa = 3.19SYY105 pKa = 11.95GSSNKK110 pKa = 9.82SSSNDD115 pKa = 2.74NDD117 pKa = 4.17TYY119 pKa = 11.56GSSNKK124 pKa = 9.56NSSDD128 pKa = 3.53TYY130 pKa = 11.12GSSNKK135 pKa = 9.55NSSDD139 pKa = 3.67SYY141 pKa = 10.85GSSGNTDD148 pKa = 2.99SYY150 pKa = 11.83GSSNKK155 pKa = 9.68NSNDD159 pKa = 3.04NDD161 pKa = 3.66DD162 pKa = 4.86SYY164 pKa = 12.21GSSNKK169 pKa = 9.79SSSNTDD175 pKa = 2.7SYY177 pKa = 11.94GSSNKK182 pKa = 9.58NSSDD186 pKa = 3.67SYY188 pKa = 10.85GSSGNTDD195 pKa = 2.99SYY197 pKa = 11.83GSSNKK202 pKa = 9.68NSNDD206 pKa = 3.2DD207 pKa = 3.71DD208 pKa = 5.25SYY210 pKa = 11.7GSSNTRR216 pKa = 11.84SSGNNNNSSDD226 pKa = 4.12SYY228 pKa = 11.61GSSNNNNNDD237 pKa = 3.11SYY239 pKa = 12.1GSGNNNSGSSTVDD252 pKa = 3.16KK253 pKa = 11.16LVDD256 pKa = 3.35TATGFLGRR264 pKa = 11.84SGGNNNNNDD273 pKa = 3.15SNYY276 pKa = 10.97
Molecular weight: 28.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B8GU12|A0A1B8GU12_9PEZI WD_REPEATS_REGION domain-containing protein OS=Pseudogymnoascus verrucosus OX=342668 GN=VE01_02574 PE=4 SV=1
MM1 pKa = 7.48SSSSPPRR8 pKa = 11.84DD9 pKa = 3.46PPPSQGGPPSIISSRR24 pKa = 11.84MTDD27 pKa = 2.8IASDD31 pKa = 4.04DD32 pKa = 3.74GHH34 pKa = 8.03DD35 pKa = 5.21LDD37 pKa = 4.67TTSHH41 pKa = 6.4ATPAHH46 pKa = 5.17RR47 pKa = 11.84TSTNRR52 pKa = 11.84QSIQSSRR59 pKa = 11.84PGTGSAAMSTRR70 pKa = 11.84SPWSASPPSRR80 pKa = 11.84RR81 pKa = 11.84GPKK84 pKa = 9.98SGGTPSILGGAAGARR99 pKa = 11.84PPSSTSRR106 pKa = 11.84THH108 pKa = 6.18VPSLTSHH115 pKa = 6.72AFFRR119 pKa = 11.84PMSSQRR125 pKa = 11.84LQAQRR130 pKa = 11.84GGPRR134 pKa = 11.84TPVAGQNQRR143 pKa = 11.84GSGDD147 pKa = 3.53FGDD150 pKa = 4.63APAVPSIASGYY161 pKa = 7.92TGNPIQSRR169 pKa = 11.84DD170 pKa = 3.52EE171 pKa = 4.39SGPPPPSRR179 pKa = 11.84GTEE182 pKa = 3.49ISVQEE187 pKa = 4.06YY188 pKa = 10.18VQTTTANTSPTHH200 pKa = 5.59GHH202 pKa = 5.87FQTTSLSSSVQPLQVNNASSNPNGLTVNTGLTTFRR237 pKa = 11.84NANLPTPSKK246 pKa = 10.14SPHH249 pKa = 5.26SFRR252 pKa = 11.84SSFLLPSRR260 pKa = 11.84GGDD263 pKa = 3.57MPPHH267 pKa = 6.11SPNRR271 pKa = 11.84SNHH274 pKa = 4.34GHH276 pKa = 6.06SKK278 pKa = 10.3LSSGATSPARR288 pKa = 11.84PSDD291 pKa = 3.83VTPLPSPLTPATGAMGEE308 pKa = 4.09KK309 pKa = 9.88HH310 pKa = 6.38GPGGNGGGGAGGGGKK325 pKa = 10.36GGVKK329 pKa = 10.25NYY331 pKa = 9.32VHH333 pKa = 6.97FPGNTRR339 pKa = 11.84FFLGGRR345 pKa = 11.84FQNARR350 pKa = 11.84DD351 pKa = 3.72RR352 pKa = 11.84PVNIATGILVVVPAVLFFVFQASWLWHH379 pKa = 5.36RR380 pKa = 11.84VSPAVPVVFAYY391 pKa = 10.78LSFICFSSFIHH402 pKa = 6.87ASVSDD407 pKa = 4.03PGILPRR413 pKa = 11.84DD414 pKa = 3.55LHH416 pKa = 6.56KK417 pKa = 10.79FPPPPATEE425 pKa = 4.85DD426 pKa = 4.11PLTLAPPTTAWLIVKK441 pKa = 9.43SHH443 pKa = 6.63LPASTAMEE451 pKa = 4.36VPVKK455 pKa = 10.49YY456 pKa = 10.6CKK458 pKa = 9.46TCHH461 pKa = 5.24IWRR464 pKa = 11.84PPRR467 pKa = 11.84GHH469 pKa = 7.0HH470 pKa = 7.04CRR472 pKa = 11.84ICDD475 pKa = 3.6NCIEE479 pKa = 4.44THH481 pKa = 6.28DD482 pKa = 4.18HH483 pKa = 5.9HH484 pKa = 7.48CVWLNNCVGRR494 pKa = 11.84RR495 pKa = 11.84NYY497 pKa = 10.45RR498 pKa = 11.84YY499 pKa = 9.85FFTFVAAGTGMAIFCIVTSVVQLSTVGRR527 pKa = 11.84DD528 pKa = 3.18NNSDD532 pKa = 3.56FGSAIPRR539 pKa = 11.84EE540 pKa = 4.06RR541 pKa = 11.84GVFALLIYY549 pKa = 10.38AALALPYY556 pKa = 9.73PAALLFYY563 pKa = 10.37HH564 pKa = 7.01IFLSGRR570 pKa = 11.84GEE572 pKa = 4.18TTRR575 pKa = 11.84EE576 pKa = 3.81LLNGRR581 pKa = 11.84KK582 pKa = 8.9FKK584 pKa = 10.62RR585 pKa = 11.84GEE587 pKa = 3.63RR588 pKa = 11.84HH589 pKa = 5.95RR590 pKa = 11.84PFTLGSVVKK599 pKa = 10.55NWIAVLGRR607 pKa = 11.84PRR609 pKa = 11.84GGGYY613 pKa = 9.66IGFKK617 pKa = 10.12RR618 pKa = 11.84VGGVWLGEE626 pKa = 4.01KK627 pKa = 10.41GVVEE631 pKa = 4.52GRR633 pKa = 11.84QGGSGGGNGEE643 pKa = 4.14MLEE646 pKa = 4.12MRR648 pKa = 11.84GMSQGAA654 pKa = 3.21
MM1 pKa = 7.48SSSSPPRR8 pKa = 11.84DD9 pKa = 3.46PPPSQGGPPSIISSRR24 pKa = 11.84MTDD27 pKa = 2.8IASDD31 pKa = 4.04DD32 pKa = 3.74GHH34 pKa = 8.03DD35 pKa = 5.21LDD37 pKa = 4.67TTSHH41 pKa = 6.4ATPAHH46 pKa = 5.17RR47 pKa = 11.84TSTNRR52 pKa = 11.84QSIQSSRR59 pKa = 11.84PGTGSAAMSTRR70 pKa = 11.84SPWSASPPSRR80 pKa = 11.84RR81 pKa = 11.84GPKK84 pKa = 9.98SGGTPSILGGAAGARR99 pKa = 11.84PPSSTSRR106 pKa = 11.84THH108 pKa = 6.18VPSLTSHH115 pKa = 6.72AFFRR119 pKa = 11.84PMSSQRR125 pKa = 11.84LQAQRR130 pKa = 11.84GGPRR134 pKa = 11.84TPVAGQNQRR143 pKa = 11.84GSGDD147 pKa = 3.53FGDD150 pKa = 4.63APAVPSIASGYY161 pKa = 7.92TGNPIQSRR169 pKa = 11.84DD170 pKa = 3.52EE171 pKa = 4.39SGPPPPSRR179 pKa = 11.84GTEE182 pKa = 3.49ISVQEE187 pKa = 4.06YY188 pKa = 10.18VQTTTANTSPTHH200 pKa = 5.59GHH202 pKa = 5.87FQTTSLSSSVQPLQVNNASSNPNGLTVNTGLTTFRR237 pKa = 11.84NANLPTPSKK246 pKa = 10.14SPHH249 pKa = 5.26SFRR252 pKa = 11.84SSFLLPSRR260 pKa = 11.84GGDD263 pKa = 3.57MPPHH267 pKa = 6.11SPNRR271 pKa = 11.84SNHH274 pKa = 4.34GHH276 pKa = 6.06SKK278 pKa = 10.3LSSGATSPARR288 pKa = 11.84PSDD291 pKa = 3.83VTPLPSPLTPATGAMGEE308 pKa = 4.09KK309 pKa = 9.88HH310 pKa = 6.38GPGGNGGGGAGGGGKK325 pKa = 10.36GGVKK329 pKa = 10.25NYY331 pKa = 9.32VHH333 pKa = 6.97FPGNTRR339 pKa = 11.84FFLGGRR345 pKa = 11.84FQNARR350 pKa = 11.84DD351 pKa = 3.72RR352 pKa = 11.84PVNIATGILVVVPAVLFFVFQASWLWHH379 pKa = 5.36RR380 pKa = 11.84VSPAVPVVFAYY391 pKa = 10.78LSFICFSSFIHH402 pKa = 6.87ASVSDD407 pKa = 4.03PGILPRR413 pKa = 11.84DD414 pKa = 3.55LHH416 pKa = 6.56KK417 pKa = 10.79FPPPPATEE425 pKa = 4.85DD426 pKa = 4.11PLTLAPPTTAWLIVKK441 pKa = 9.43SHH443 pKa = 6.63LPASTAMEE451 pKa = 4.36VPVKK455 pKa = 10.49YY456 pKa = 10.6CKK458 pKa = 9.46TCHH461 pKa = 5.24IWRR464 pKa = 11.84PPRR467 pKa = 11.84GHH469 pKa = 7.0HH470 pKa = 7.04CRR472 pKa = 11.84ICDD475 pKa = 3.6NCIEE479 pKa = 4.44THH481 pKa = 6.28DD482 pKa = 4.18HH483 pKa = 5.9HH484 pKa = 7.48CVWLNNCVGRR494 pKa = 11.84RR495 pKa = 11.84NYY497 pKa = 10.45RR498 pKa = 11.84YY499 pKa = 9.85FFTFVAAGTGMAIFCIVTSVVQLSTVGRR527 pKa = 11.84DD528 pKa = 3.18NNSDD532 pKa = 3.56FGSAIPRR539 pKa = 11.84EE540 pKa = 4.06RR541 pKa = 11.84GVFALLIYY549 pKa = 10.38AALALPYY556 pKa = 9.73PAALLFYY563 pKa = 10.37HH564 pKa = 7.01IFLSGRR570 pKa = 11.84GEE572 pKa = 4.18TTRR575 pKa = 11.84EE576 pKa = 3.81LLNGRR581 pKa = 11.84KK582 pKa = 8.9FKK584 pKa = 10.62RR585 pKa = 11.84GEE587 pKa = 3.63RR588 pKa = 11.84HH589 pKa = 5.95RR590 pKa = 11.84PFTLGSVVKK599 pKa = 10.55NWIAVLGRR607 pKa = 11.84PRR609 pKa = 11.84GGGYY613 pKa = 9.66IGFKK617 pKa = 10.12RR618 pKa = 11.84VGGVWLGEE626 pKa = 4.01KK627 pKa = 10.41GVVEE631 pKa = 4.52GRR633 pKa = 11.84QGGSGGGNGEE643 pKa = 4.14MLEE646 pKa = 4.12MRR648 pKa = 11.84GMSQGAA654 pKa = 3.21
Molecular weight: 69.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5291759 |
51 |
5681 |
495.3 |
54.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.92 ± 0.023 | 1.194 ± 0.009 |
5.561 ± 0.02 | 6.269 ± 0.026 |
3.681 ± 0.015 | 7.342 ± 0.026 |
2.211 ± 0.01 | 5.061 ± 0.017 |
4.956 ± 0.023 | 8.74 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.205 ± 0.009 | 3.716 ± 0.012 |
5.957 ± 0.026 | 3.796 ± 0.02 |
5.81 ± 0.019 | 8.08 ± 0.024 |
6.081 ± 0.019 | 6.205 ± 0.017 |
1.433 ± 0.008 | 2.78 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
