
Marinilabilia rubra
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Marinilabiliaceae; Marinilabilia
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
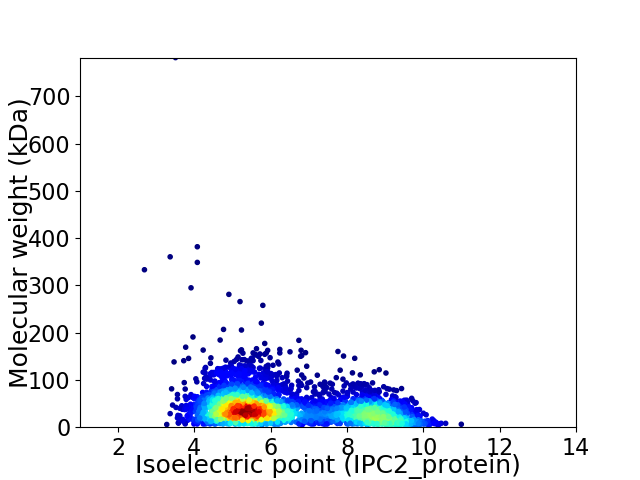
Virtual 2D-PAGE plot for 4010 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
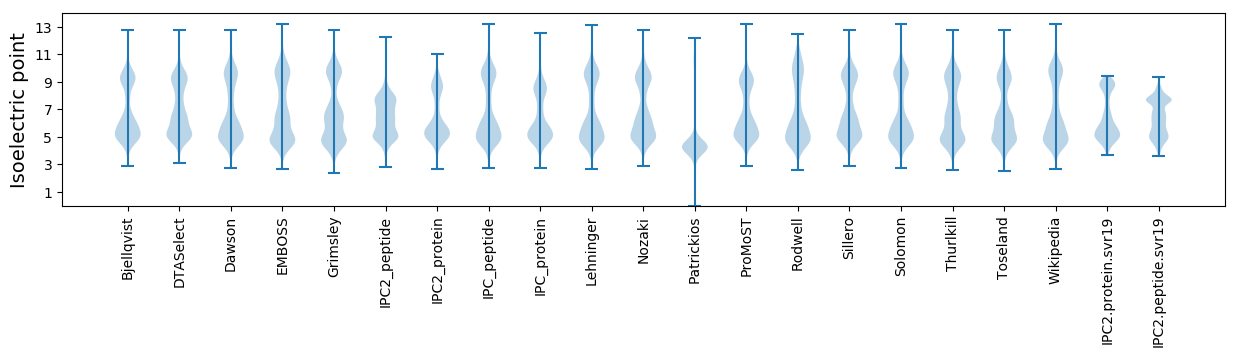
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U2BBP0|A0A2U2BBP0_9BACT Site-specific DNA-methyltransferase (adenine-specific) OS=Marinilabilia rubra OX=2162893 GN=DDZ16_06080 PE=3 SV=1
MM1 pKa = 7.12QLSKK5 pKa = 9.41TWWYY9 pKa = 11.01FLMATIFVFTACDD22 pKa = 4.14DD23 pKa = 4.59DD24 pKa = 7.72DD25 pKa = 7.24DD26 pKa = 7.44DD27 pKa = 6.2EE28 pKa = 5.73DD29 pKa = 6.27LNGNWVEE36 pKa = 4.03LGAFEE41 pKa = 5.67GIPRR45 pKa = 11.84TDD47 pKa = 2.65AVAFVIGDD55 pKa = 3.55YY56 pKa = 10.73AYY58 pKa = 11.22VGTGYY63 pKa = 10.65NGEE66 pKa = 3.91EE67 pKa = 4.23DD68 pKa = 3.34EE69 pKa = 4.75RR70 pKa = 11.84LRR72 pKa = 11.84DD73 pKa = 3.91FWRR76 pKa = 11.84YY77 pKa = 9.89DD78 pKa = 3.23AVNDD82 pKa = 3.38TWRR85 pKa = 11.84EE86 pKa = 4.03VASLPAEE93 pKa = 3.86AAARR97 pKa = 11.84NGAVAFSSNGKK108 pKa = 9.81GYY110 pKa = 10.93VGTGYY115 pKa = 10.79DD116 pKa = 4.27GEE118 pKa = 4.68NKK120 pKa = 10.51LKK122 pKa = 10.76DD123 pKa = 3.56FWEE126 pKa = 4.62FNPAEE131 pKa = 4.6GGDD134 pKa = 4.16GQWTRR139 pKa = 11.84IDD141 pKa = 4.68DD142 pKa = 4.14FPGTARR148 pKa = 11.84YY149 pKa = 9.0SAIAFGLEE157 pKa = 3.5QNGYY161 pKa = 10.21VGTGYY166 pKa = 10.82DD167 pKa = 3.74GNRR170 pKa = 11.84LKK172 pKa = 11.0DD173 pKa = 3.96FYY175 pKa = 11.17KK176 pKa = 10.69YY177 pKa = 10.59DD178 pKa = 3.78AASGSWSQIMSIGGSKK194 pKa = 10.21RR195 pKa = 11.84RR196 pKa = 11.84DD197 pKa = 3.09AAVFVIDD204 pKa = 3.87GKK206 pKa = 11.09AYY208 pKa = 10.81VFTGVDD214 pKa = 2.97NSEE217 pKa = 4.36YY218 pKa = 10.05VTDD221 pKa = 3.06AWMYY225 pKa = 11.01DD226 pKa = 3.57PQADD230 pKa = 3.12TWTEE234 pKa = 3.59KK235 pKa = 11.07RR236 pKa = 11.84EE237 pKa = 4.18ITSGTNDD244 pKa = 3.75DD245 pKa = 4.1EE246 pKa = 6.43SYY248 pKa = 11.78DD249 pKa = 4.11DD250 pKa = 5.36DD251 pKa = 4.9YY252 pKa = 12.16SLAGTNVVAFSMGGLGYY269 pKa = 10.08ISTGGQGYY277 pKa = 10.47AGNTTWEE284 pKa = 4.35YY285 pKa = 11.77NPITDD290 pKa = 3.79FWEE293 pKa = 4.36EE294 pKa = 3.6KK295 pKa = 10.82SNFEE299 pKa = 4.03GSSRR303 pKa = 11.84SDD305 pKa = 2.86AVAFAINNTPYY316 pKa = 10.67VATGNSSGYY325 pKa = 10.63YY326 pKa = 10.02FDD328 pKa = 5.98DD329 pKa = 2.86VWTFYY334 pKa = 10.57PDD336 pKa = 3.81EE337 pKa = 4.31EE338 pKa = 5.08QNDD341 pKa = 3.38NDD343 pKa = 3.73
MM1 pKa = 7.12QLSKK5 pKa = 9.41TWWYY9 pKa = 11.01FLMATIFVFTACDD22 pKa = 4.14DD23 pKa = 4.59DD24 pKa = 7.72DD25 pKa = 7.24DD26 pKa = 7.44DD27 pKa = 6.2EE28 pKa = 5.73DD29 pKa = 6.27LNGNWVEE36 pKa = 4.03LGAFEE41 pKa = 5.67GIPRR45 pKa = 11.84TDD47 pKa = 2.65AVAFVIGDD55 pKa = 3.55YY56 pKa = 10.73AYY58 pKa = 11.22VGTGYY63 pKa = 10.65NGEE66 pKa = 3.91EE67 pKa = 4.23DD68 pKa = 3.34EE69 pKa = 4.75RR70 pKa = 11.84LRR72 pKa = 11.84DD73 pKa = 3.91FWRR76 pKa = 11.84YY77 pKa = 9.89DD78 pKa = 3.23AVNDD82 pKa = 3.38TWRR85 pKa = 11.84EE86 pKa = 4.03VASLPAEE93 pKa = 3.86AAARR97 pKa = 11.84NGAVAFSSNGKK108 pKa = 9.81GYY110 pKa = 10.93VGTGYY115 pKa = 10.79DD116 pKa = 4.27GEE118 pKa = 4.68NKK120 pKa = 10.51LKK122 pKa = 10.76DD123 pKa = 3.56FWEE126 pKa = 4.62FNPAEE131 pKa = 4.6GGDD134 pKa = 4.16GQWTRR139 pKa = 11.84IDD141 pKa = 4.68DD142 pKa = 4.14FPGTARR148 pKa = 11.84YY149 pKa = 9.0SAIAFGLEE157 pKa = 3.5QNGYY161 pKa = 10.21VGTGYY166 pKa = 10.82DD167 pKa = 3.74GNRR170 pKa = 11.84LKK172 pKa = 11.0DD173 pKa = 3.96FYY175 pKa = 11.17KK176 pKa = 10.69YY177 pKa = 10.59DD178 pKa = 3.78AASGSWSQIMSIGGSKK194 pKa = 10.21RR195 pKa = 11.84RR196 pKa = 11.84DD197 pKa = 3.09AAVFVIDD204 pKa = 3.87GKK206 pKa = 11.09AYY208 pKa = 10.81VFTGVDD214 pKa = 2.97NSEE217 pKa = 4.36YY218 pKa = 10.05VTDD221 pKa = 3.06AWMYY225 pKa = 11.01DD226 pKa = 3.57PQADD230 pKa = 3.12TWTEE234 pKa = 3.59KK235 pKa = 11.07RR236 pKa = 11.84EE237 pKa = 4.18ITSGTNDD244 pKa = 3.75DD245 pKa = 4.1EE246 pKa = 6.43SYY248 pKa = 11.78DD249 pKa = 4.11DD250 pKa = 5.36DD251 pKa = 4.9YY252 pKa = 12.16SLAGTNVVAFSMGGLGYY269 pKa = 10.08ISTGGQGYY277 pKa = 10.47AGNTTWEE284 pKa = 4.35YY285 pKa = 11.77NPITDD290 pKa = 3.79FWEE293 pKa = 4.36EE294 pKa = 3.6KK295 pKa = 10.82SNFEE299 pKa = 4.03GSSRR303 pKa = 11.84SDD305 pKa = 2.86AVAFAINNTPYY316 pKa = 10.67VATGNSSGYY325 pKa = 10.63YY326 pKa = 10.02FDD328 pKa = 5.98DD329 pKa = 2.86VWTFYY334 pKa = 10.57PDD336 pKa = 3.81EE337 pKa = 4.31EE338 pKa = 5.08QNDD341 pKa = 3.38NDD343 pKa = 3.73
Molecular weight: 38.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U2B8G7|A0A2U2B8G7_9BACT Uncharacterized protein OS=Marinilabilia rubra OX=2162893 GN=DDZ16_10130 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.98HH16 pKa = 3.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MTTVNGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.54GRR39 pKa = 11.84KK40 pKa = 8.81RR41 pKa = 11.84LTVSNEE47 pKa = 3.61KK48 pKa = 9.93RR49 pKa = 11.84HH50 pKa = 5.47KK51 pKa = 10.53AA52 pKa = 3.32
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.98HH16 pKa = 3.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MTTVNGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.54GRR39 pKa = 11.84KK40 pKa = 8.81RR41 pKa = 11.84LTVSNEE47 pKa = 3.61KK48 pKa = 9.93RR49 pKa = 11.84HH50 pKa = 5.47KK51 pKa = 10.53AA52 pKa = 3.32
Molecular weight: 6.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1511230 |
27 |
7413 |
376.9 |
42.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.309 ± 0.032 | 0.76 ± 0.012 |
5.755 ± 0.031 | 7.062 ± 0.036 |
5.159 ± 0.033 | 7.042 ± 0.043 |
1.946 ± 0.019 | 6.976 ± 0.036 |
6.746 ± 0.054 | 9.24 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.022 | 5.457 ± 0.036 |
3.919 ± 0.021 | 3.41 ± 0.021 |
4.304 ± 0.029 | 6.684 ± 0.04 |
5.146 ± 0.049 | 6.462 ± 0.032 |
1.286 ± 0.015 | 3.868 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
