
Pseudomonas jinjuensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas aeruginosa group
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
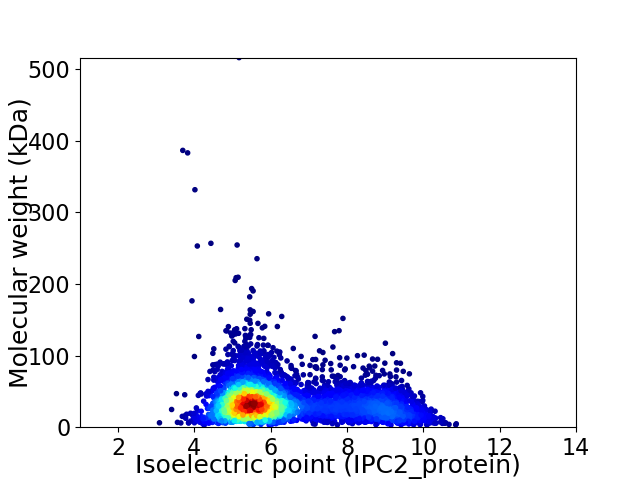
Virtual 2D-PAGE plot for 5169 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0BER0|A0A1H0BER0_9PSED Uncharacterized protein OS=Pseudomonas jinjuensis OX=198616 GN=SAMN05216193_10342 PE=4 SV=1
MM1 pKa = 7.48FKK3 pKa = 10.77LKK5 pKa = 10.14PLAIAILTLASAQVMAAGNTATQIQLSGDD34 pKa = 4.12GNDD37 pKa = 3.41STAMQTGISDD47 pKa = 4.27GNTSTQVQAGDD58 pKa = 3.48NNTANVRR65 pKa = 11.84QLDD68 pKa = 3.77VSTGNVAGQAQAGNGNTATIEE89 pKa = 4.35QKK91 pKa = 9.63WGAYY95 pKa = 8.76YY96 pKa = 10.94NEE98 pKa = 4.33AAQAQTGNGNTATVDD113 pKa = 3.34QHH115 pKa = 8.43DD116 pKa = 4.42EE117 pKa = 3.73ATANHH122 pKa = 6.56VYY124 pKa = 9.94QVQDD128 pKa = 3.26GSANKK133 pKa = 10.14AFAEE137 pKa = 4.02NAYY140 pKa = 10.6VSTGNYY146 pKa = 7.38ATQLQSGTSHH156 pKa = 6.4EE157 pKa = 4.11AEE159 pKa = 4.34IWQQWGEE166 pKa = 4.21GNQALQVQDD175 pKa = 4.63GDD177 pKa = 3.54DD178 pKa = 4.24HH179 pKa = 5.83YY180 pKa = 12.14AKK182 pKa = 10.41VYY184 pKa = 10.42QGADD188 pKa = 3.15ANSNISVQTQTGTSNTSTVNQLFSADD214 pKa = 3.49GNYY217 pKa = 10.3SEE219 pKa = 4.63VTQNGVGAGNSALVEE234 pKa = 4.24QNWGGGNVSVLDD246 pKa = 3.71QDD248 pKa = 4.59GDD250 pKa = 3.97GNDD253 pKa = 3.11AHH255 pKa = 6.48VTQQHH260 pKa = 6.12GDD262 pKa = 3.63GNGSALTQAGNGNLASVSQHH282 pKa = 6.96DD283 pKa = 3.85GDD285 pKa = 4.72GNQSALTQTGNYY297 pKa = 9.26NSASVTQADD306 pKa = 4.04GGWNVSSLSQSGNNHH321 pKa = 5.5QATVTQGGGNSNEE334 pKa = 4.63LYY336 pKa = 10.67FEE338 pKa = 4.07QDD340 pKa = 2.71DD341 pKa = 4.39DD342 pKa = 5.7GNVLTADD349 pKa = 3.85QSGTDD354 pKa = 3.68NLITGYY360 pKa = 9.97SHH362 pKa = 7.57GDD364 pKa = 3.55DD365 pKa = 3.3NVVDD369 pKa = 3.92VDD371 pKa = 4.38QVGSLNTAHH380 pKa = 6.42VEE382 pKa = 4.17QYY384 pKa = 10.74SGSSEE389 pKa = 3.87NLADD393 pKa = 3.83ISQTGSDD400 pKa = 3.35HH401 pKa = 6.35VATVLQGGMGNEE413 pKa = 3.84AFVTQTSTGNTAYY426 pKa = 10.8VLQAGTNGTATIIQNN441 pKa = 3.44
MM1 pKa = 7.48FKK3 pKa = 10.77LKK5 pKa = 10.14PLAIAILTLASAQVMAAGNTATQIQLSGDD34 pKa = 4.12GNDD37 pKa = 3.41STAMQTGISDD47 pKa = 4.27GNTSTQVQAGDD58 pKa = 3.48NNTANVRR65 pKa = 11.84QLDD68 pKa = 3.77VSTGNVAGQAQAGNGNTATIEE89 pKa = 4.35QKK91 pKa = 9.63WGAYY95 pKa = 8.76YY96 pKa = 10.94NEE98 pKa = 4.33AAQAQTGNGNTATVDD113 pKa = 3.34QHH115 pKa = 8.43DD116 pKa = 4.42EE117 pKa = 3.73ATANHH122 pKa = 6.56VYY124 pKa = 9.94QVQDD128 pKa = 3.26GSANKK133 pKa = 10.14AFAEE137 pKa = 4.02NAYY140 pKa = 10.6VSTGNYY146 pKa = 7.38ATQLQSGTSHH156 pKa = 6.4EE157 pKa = 4.11AEE159 pKa = 4.34IWQQWGEE166 pKa = 4.21GNQALQVQDD175 pKa = 4.63GDD177 pKa = 3.54DD178 pKa = 4.24HH179 pKa = 5.83YY180 pKa = 12.14AKK182 pKa = 10.41VYY184 pKa = 10.42QGADD188 pKa = 3.15ANSNISVQTQTGTSNTSTVNQLFSADD214 pKa = 3.49GNYY217 pKa = 10.3SEE219 pKa = 4.63VTQNGVGAGNSALVEE234 pKa = 4.24QNWGGGNVSVLDD246 pKa = 3.71QDD248 pKa = 4.59GDD250 pKa = 3.97GNDD253 pKa = 3.11AHH255 pKa = 6.48VTQQHH260 pKa = 6.12GDD262 pKa = 3.63GNGSALTQAGNGNLASVSQHH282 pKa = 6.96DD283 pKa = 3.85GDD285 pKa = 4.72GNQSALTQTGNYY297 pKa = 9.26NSASVTQADD306 pKa = 4.04GGWNVSSLSQSGNNHH321 pKa = 5.5QATVTQGGGNSNEE334 pKa = 4.63LYY336 pKa = 10.67FEE338 pKa = 4.07QDD340 pKa = 2.71DD341 pKa = 4.39DD342 pKa = 5.7GNVLTADD349 pKa = 3.85QSGTDD354 pKa = 3.68NLITGYY360 pKa = 9.97SHH362 pKa = 7.57GDD364 pKa = 3.55DD365 pKa = 3.3NVVDD369 pKa = 3.92VDD371 pKa = 4.38QVGSLNTAHH380 pKa = 6.42VEE382 pKa = 4.17QYY384 pKa = 10.74SGSSEE389 pKa = 3.87NLADD393 pKa = 3.83ISQTGSDD400 pKa = 3.35HH401 pKa = 6.35VATVLQGGMGNEE413 pKa = 3.84AFVTQTSTGNTAYY426 pKa = 10.8VLQAGTNGTATIIQNN441 pKa = 3.44
Molecular weight: 45.31 kDa
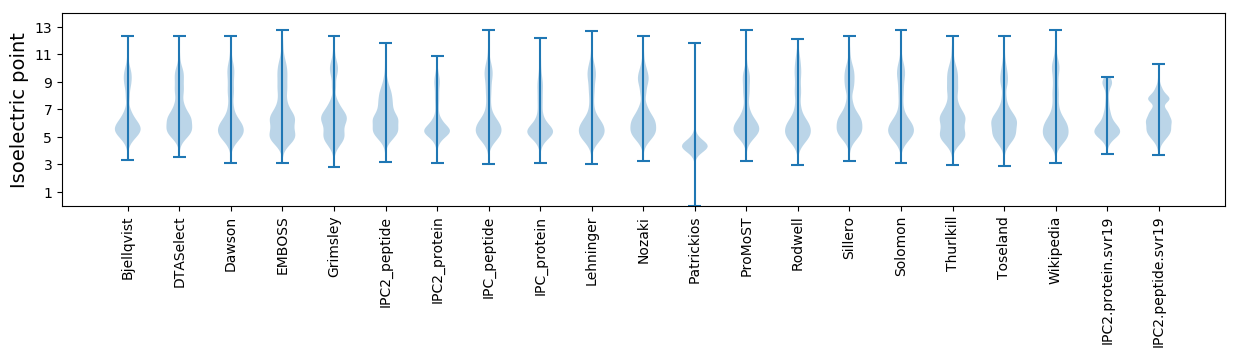
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0JA05|A0A1H0JA05_9PSED Uncharacterized protein YydD contains DUF2326 domain OS=Pseudomonas jinjuensis OX=198616 GN=SAMN05216193_1114 PE=4 SV=1
MM1 pKa = 7.55PRR3 pKa = 11.84YY4 pKa = 10.08ALITGASSGIGLALAEE20 pKa = 4.06ALARR24 pKa = 11.84RR25 pKa = 11.84GQALVLVARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84DD37 pKa = 3.63ALEE40 pKa = 4.64SIACEE45 pKa = 3.49LAQRR49 pKa = 11.84FGVDD53 pKa = 3.18VLFRR57 pKa = 11.84TCDD60 pKa = 3.28LSEE63 pKa = 4.27PLQISGLLHH72 pKa = 6.66EE73 pKa = 5.77LDD75 pKa = 3.99QNGLDD80 pKa = 4.14IEE82 pKa = 4.63LLVNSAGIGSAGAFLEE98 pKa = 4.56HH99 pKa = 6.95DD100 pKa = 3.45WSRR103 pKa = 11.84EE104 pKa = 3.81RR105 pKa = 11.84EE106 pKa = 4.18LVEE109 pKa = 4.84LNVLALTRR117 pKa = 11.84LCHH120 pKa = 7.24AIGNRR125 pKa = 11.84MAQNGGGRR133 pKa = 11.84ILNVASLAALHH144 pKa = 6.49PGPWMSSYY152 pKa = 10.19HH153 pKa = 5.95ASKK156 pKa = 10.72AYY158 pKa = 10.08VLSFSEE164 pKa = 4.4GLRR167 pKa = 11.84EE168 pKa = 3.89EE169 pKa = 4.23LKK171 pKa = 10.79GRR173 pKa = 11.84GVKK176 pKa = 10.31VSLLCPGPTRR186 pKa = 11.84SAFFRR191 pKa = 11.84HH192 pKa = 6.13AGLDD196 pKa = 3.55GEE198 pKa = 4.37RR199 pKa = 11.84FEE201 pKa = 4.77GKK203 pKa = 10.28RR204 pKa = 11.84MLSPEE209 pKa = 3.64QVAFHH214 pKa = 5.94TVRR217 pKa = 11.84ALQRR221 pKa = 11.84NPAIIIPGWRR231 pKa = 11.84NRR233 pKa = 11.84LLAWCPRR240 pKa = 11.84LAPRR244 pKa = 11.84WLVRR248 pKa = 11.84KK249 pKa = 9.21LAGRR253 pKa = 11.84RR254 pKa = 11.84NRR256 pKa = 11.84LAVQRR261 pKa = 4.27
MM1 pKa = 7.55PRR3 pKa = 11.84YY4 pKa = 10.08ALITGASSGIGLALAEE20 pKa = 4.06ALARR24 pKa = 11.84RR25 pKa = 11.84GQALVLVARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84DD37 pKa = 3.63ALEE40 pKa = 4.64SIACEE45 pKa = 3.49LAQRR49 pKa = 11.84FGVDD53 pKa = 3.18VLFRR57 pKa = 11.84TCDD60 pKa = 3.28LSEE63 pKa = 4.27PLQISGLLHH72 pKa = 6.66EE73 pKa = 5.77LDD75 pKa = 3.99QNGLDD80 pKa = 4.14IEE82 pKa = 4.63LLVNSAGIGSAGAFLEE98 pKa = 4.56HH99 pKa = 6.95DD100 pKa = 3.45WSRR103 pKa = 11.84EE104 pKa = 3.81RR105 pKa = 11.84EE106 pKa = 4.18LVEE109 pKa = 4.84LNVLALTRR117 pKa = 11.84LCHH120 pKa = 7.24AIGNRR125 pKa = 11.84MAQNGGGRR133 pKa = 11.84ILNVASLAALHH144 pKa = 6.49PGPWMSSYY152 pKa = 10.19HH153 pKa = 5.95ASKK156 pKa = 10.72AYY158 pKa = 10.08VLSFSEE164 pKa = 4.4GLRR167 pKa = 11.84EE168 pKa = 3.89EE169 pKa = 4.23LKK171 pKa = 10.79GRR173 pKa = 11.84GVKK176 pKa = 10.31VSLLCPGPTRR186 pKa = 11.84SAFFRR191 pKa = 11.84HH192 pKa = 6.13AGLDD196 pKa = 3.55GEE198 pKa = 4.37RR199 pKa = 11.84FEE201 pKa = 4.77GKK203 pKa = 10.28RR204 pKa = 11.84MLSPEE209 pKa = 3.64QVAFHH214 pKa = 5.94TVRR217 pKa = 11.84ALQRR221 pKa = 11.84NPAIIIPGWRR231 pKa = 11.84NRR233 pKa = 11.84LLAWCPRR240 pKa = 11.84LAPRR244 pKa = 11.84WLVRR248 pKa = 11.84KK249 pKa = 9.21LAGRR253 pKa = 11.84RR254 pKa = 11.84NRR256 pKa = 11.84LAVQRR261 pKa = 4.27
Molecular weight: 28.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1739259 |
26 |
4699 |
336.5 |
36.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.35 ± 0.039 | 1.042 ± 0.013 |
5.468 ± 0.027 | 6.092 ± 0.033 |
3.618 ± 0.021 | 8.444 ± 0.036 |
2.251 ± 0.016 | 4.54 ± 0.024 |
3.024 ± 0.029 | 11.756 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.272 ± 0.019 | 2.796 ± 0.026 |
5.031 ± 0.027 | 4.229 ± 0.023 |
7.206 ± 0.037 | 5.471 ± 0.021 |
4.464 ± 0.03 | 6.985 ± 0.032 |
1.447 ± 0.016 | 2.516 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
