
Pneumocystis murina (strain B123) (Mouse pneumocystis pneumonia agent) (Pneumocystis carinii f. sp. muris)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; Taphrinomycotina; Pneumocystidomycetes; Pneumocystidales; Pneumocystidaceae; Pneumocystis; Pneumocystis murina
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3635 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
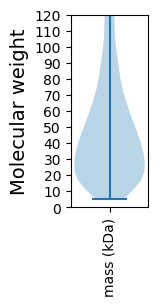
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|M7NRW1|M7NRW1_PNEMU Uncharacterized protein OS=Pneumocystis murina (strain B123) OX=1069680 GN=PNEG_01772 PE=4 SV=1
MM1 pKa = 7.52GFLFVTFLFAILTLINAYY19 pKa = 9.34KK20 pKa = 10.65VLLPKK25 pKa = 10.54PGDD28 pKa = 3.01KK29 pKa = 10.07WEE31 pKa = 4.98PGLHH35 pKa = 4.55QVTWEE40 pKa = 4.09AVDD43 pKa = 3.62DD44 pKa = 4.14TSTVNVYY51 pKa = 8.07LHH53 pKa = 7.09RR54 pKa = 11.84NNDD57 pKa = 3.29NPPFYY62 pKa = 10.87LLLAAKK68 pKa = 10.31VPLKK72 pKa = 10.57DD73 pKa = 3.53GKK75 pKa = 10.9ADD77 pKa = 3.45VPIPYY82 pKa = 9.95NVIPGSEE89 pKa = 4.12YY90 pKa = 10.69TILLTSLNPYY100 pKa = 10.12DD101 pKa = 4.43VYY103 pKa = 10.78TSSGGFEE110 pKa = 3.54ITEE113 pKa = 4.4AKK115 pKa = 10.5DD116 pKa = 3.39GLKK119 pKa = 10.36GVCVDD124 pKa = 3.64YY125 pKa = 11.47LKK127 pKa = 11.01DD128 pKa = 4.43DD129 pKa = 4.12GSSTIGPIPGDD140 pKa = 3.63TRR142 pKa = 11.84PVTTSLLPPSPTPPSPAPPAPTPAPPAPTPPSPEE176 pKa = 4.04PPAPAPPEE184 pKa = 4.34GEE186 pKa = 4.01VEE188 pKa = 4.15NCDD191 pKa = 3.34EE192 pKa = 4.5CDD194 pKa = 3.18EE195 pKa = 4.69CEE197 pKa = 4.11EE198 pKa = 4.75CGEE201 pKa = 4.32CEE203 pKa = 4.04PGEE206 pKa = 4.65GCDD209 pKa = 5.54CDD211 pKa = 5.89ADD213 pKa = 4.04DD214 pKa = 6.09HH215 pKa = 8.3GDD217 pKa = 3.56DD218 pKa = 6.06DD219 pKa = 4.57YY220 pKa = 12.16DD221 pKa = 4.13KK222 pKa = 11.67DD223 pKa = 3.7EE224 pKa = 4.7DD225 pKa = 4.34HH226 pKa = 6.7GHH228 pKa = 5.79SHH230 pKa = 8.01DD231 pKa = 6.22DD232 pKa = 5.39DD233 pKa = 7.24DD234 pKa = 7.67DD235 pKa = 7.27DD236 pKa = 7.59DD237 pKa = 6.37DD238 pKa = 5.42DD239 pKa = 6.07HH240 pKa = 7.82EE241 pKa = 5.24NDD243 pKa = 5.25HH244 pKa = 7.68DD245 pKa = 6.71DD246 pKa = 5.89DD247 pKa = 7.59DD248 pKa = 7.37DD249 pKa = 7.3DD250 pKa = 6.42DD251 pKa = 4.86DD252 pKa = 5.67HH253 pKa = 7.29GHH255 pKa = 6.04SHH257 pKa = 8.03DD258 pKa = 5.63DD259 pKa = 3.93DD260 pKa = 7.02DD261 pKa = 6.22DD262 pKa = 4.41GHH264 pKa = 7.62DD265 pKa = 4.22HH266 pKa = 7.45GEE268 pKa = 4.48DD269 pKa = 3.35KK270 pKa = 11.38NKK272 pKa = 10.03LANIGSTNIVTRR284 pKa = 11.84FWLTMMATISTILFLL299 pKa = 5.42
MM1 pKa = 7.52GFLFVTFLFAILTLINAYY19 pKa = 9.34KK20 pKa = 10.65VLLPKK25 pKa = 10.54PGDD28 pKa = 3.01KK29 pKa = 10.07WEE31 pKa = 4.98PGLHH35 pKa = 4.55QVTWEE40 pKa = 4.09AVDD43 pKa = 3.62DD44 pKa = 4.14TSTVNVYY51 pKa = 8.07LHH53 pKa = 7.09RR54 pKa = 11.84NNDD57 pKa = 3.29NPPFYY62 pKa = 10.87LLLAAKK68 pKa = 10.31VPLKK72 pKa = 10.57DD73 pKa = 3.53GKK75 pKa = 10.9ADD77 pKa = 3.45VPIPYY82 pKa = 9.95NVIPGSEE89 pKa = 4.12YY90 pKa = 10.69TILLTSLNPYY100 pKa = 10.12DD101 pKa = 4.43VYY103 pKa = 10.78TSSGGFEE110 pKa = 3.54ITEE113 pKa = 4.4AKK115 pKa = 10.5DD116 pKa = 3.39GLKK119 pKa = 10.36GVCVDD124 pKa = 3.64YY125 pKa = 11.47LKK127 pKa = 11.01DD128 pKa = 4.43DD129 pKa = 4.12GSSTIGPIPGDD140 pKa = 3.63TRR142 pKa = 11.84PVTTSLLPPSPTPPSPAPPAPTPAPPAPTPPSPEE176 pKa = 4.04PPAPAPPEE184 pKa = 4.34GEE186 pKa = 4.01VEE188 pKa = 4.15NCDD191 pKa = 3.34EE192 pKa = 4.5CDD194 pKa = 3.18EE195 pKa = 4.69CEE197 pKa = 4.11EE198 pKa = 4.75CGEE201 pKa = 4.32CEE203 pKa = 4.04PGEE206 pKa = 4.65GCDD209 pKa = 5.54CDD211 pKa = 5.89ADD213 pKa = 4.04DD214 pKa = 6.09HH215 pKa = 8.3GDD217 pKa = 3.56DD218 pKa = 6.06DD219 pKa = 4.57YY220 pKa = 12.16DD221 pKa = 4.13KK222 pKa = 11.67DD223 pKa = 3.7EE224 pKa = 4.7DD225 pKa = 4.34HH226 pKa = 6.7GHH228 pKa = 5.79SHH230 pKa = 8.01DD231 pKa = 6.22DD232 pKa = 5.39DD233 pKa = 7.24DD234 pKa = 7.67DD235 pKa = 7.27DD236 pKa = 7.59DD237 pKa = 6.37DD238 pKa = 5.42DD239 pKa = 6.07HH240 pKa = 7.82EE241 pKa = 5.24NDD243 pKa = 5.25HH244 pKa = 7.68DD245 pKa = 6.71DD246 pKa = 5.89DD247 pKa = 7.59DD248 pKa = 7.37DD249 pKa = 7.3DD250 pKa = 6.42DD251 pKa = 4.86DD252 pKa = 5.67HH253 pKa = 7.29GHH255 pKa = 6.04SHH257 pKa = 8.03DD258 pKa = 5.63DD259 pKa = 3.93DD260 pKa = 7.02DD261 pKa = 6.22DD262 pKa = 4.41GHH264 pKa = 7.62DD265 pKa = 4.22HH266 pKa = 7.45GEE268 pKa = 4.48DD269 pKa = 3.35KK270 pKa = 11.38NKK272 pKa = 10.03LANIGSTNIVTRR284 pKa = 11.84FWLTMMATISTILFLL299 pKa = 5.42
Molecular weight: 32.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7P8Q4|M7P8Q4_PNEMU Protein kinase domain-containing protein OS=Pneumocystis murina (strain B123) OX=1069680 GN=PNEG_01497 PE=4 SV=1
MM1 pKa = 7.57SSHH4 pKa = 5.19KK5 pKa = 8.98TFRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.78IILARR15 pKa = 11.84AQRR18 pKa = 11.84QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNRR34 pKa = 11.84IRR36 pKa = 11.84YY37 pKa = 5.66NAKK40 pKa = 8.4RR41 pKa = 11.84KK42 pKa = 8.21HH43 pKa = 4.15WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 11.1LNII51 pKa = 3.83
MM1 pKa = 7.57SSHH4 pKa = 5.19KK5 pKa = 8.98TFRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.78IILARR15 pKa = 11.84AQRR18 pKa = 11.84QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNRR34 pKa = 11.84IRR36 pKa = 11.84YY37 pKa = 5.66NAKK40 pKa = 8.4RR41 pKa = 11.84KK42 pKa = 8.21HH43 pKa = 4.15WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 11.1LNII51 pKa = 3.83
Molecular weight: 6.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1664453 |
45 |
5058 |
457.9 |
52.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.177 ± 0.031 | 1.62 ± 0.024 |
5.419 ± 0.025 | 7.017 ± 0.04 |
4.914 ± 0.029 | 4.272 ± 0.038 |
2.157 ± 0.017 | 8.665 ± 0.045 |
8.532 ± 0.047 | 9.921 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.076 ± 0.014 | 6.239 ± 0.038 |
3.632 ± 0.027 | 3.593 ± 0.022 |
4.413 ± 0.026 | 8.713 ± 0.047 |
5.046 ± 0.028 | 4.741 ± 0.029 |
1.067 ± 0.012 | 3.787 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
