
Gluconacetobacter liquefaciens (Acetobacter liquefaciens)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Gluconacetobacter
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
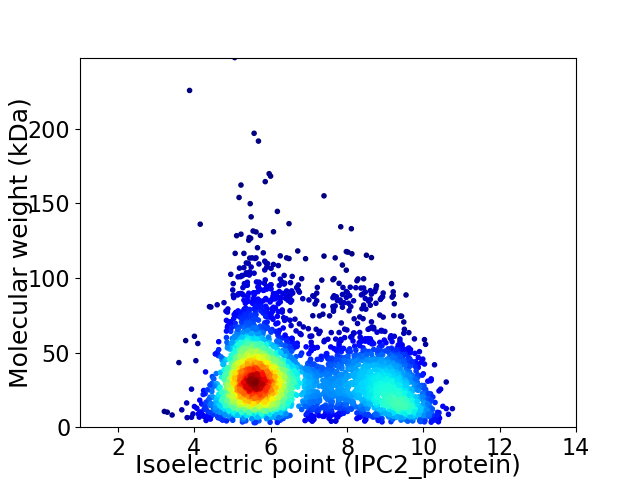
Virtual 2D-PAGE plot for 3696 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370G705|A0A370G705_GLULI L-serine dehydratase OS=Gluconacetobacter liquefaciens OX=89584 GN=C7453_102369 PE=3 SV=1
MM1 pKa = 7.41SVFNALTTAVSGINAQSTAFTNLSNNIANSQTVGYY36 pKa = 9.73KK37 pKa = 9.3ATTTAFQDD45 pKa = 4.32FVAGSLGKK53 pKa = 9.75NASSADD59 pKa = 3.27ISDD62 pKa = 3.47SVAAVTVQHH71 pKa = 6.58VDD73 pKa = 3.32NQGTAATSTDD83 pKa = 3.4SLATAISGSGLFVVSKK99 pKa = 10.61EE100 pKa = 4.29SGASTSTNTQFQSQNYY116 pKa = 4.93YY117 pKa = 8.09TRR119 pKa = 11.84NGEE122 pKa = 4.18VYY124 pKa = 10.28KK125 pKa = 11.11NNSGYY130 pKa = 10.79LVNTSGYY137 pKa = 9.17YY138 pKa = 10.07LDD140 pKa = 5.48GYY142 pKa = 9.95MVDD145 pKa = 3.85PTTGSLNASLTQLNVANVAFRR166 pKa = 11.84PTQTTTLTLSATVGTMPSSNASYY189 pKa = 8.96TAADD193 pKa = 3.57AQSYY197 pKa = 6.01TTAPVTTYY205 pKa = 10.67DD206 pKa = 4.74ASATPHH212 pKa = 6.76KK213 pKa = 10.27IALSWTQSGTNPLVWNVSAYY233 pKa = 10.49DD234 pKa = 3.66ADD236 pKa = 4.06GTGTIASNSYY246 pKa = 10.25QVTFDD251 pKa = 3.58SSGSLASVADD261 pKa = 3.63STTGKK266 pKa = 9.05TIGSTLTGASASIPITADD284 pKa = 3.28YY285 pKa = 11.22NGVAQNITLDD295 pKa = 3.52LGTIGGTSGTVMAASTGTASTNQANALTASGTTLTMSKK333 pKa = 10.43SDD335 pKa = 3.97VIGTTTGSNQTSMTSPTVANGTSVAAKK362 pKa = 6.92WTQTAASPPTWSLSLVDD379 pKa = 5.33PYY381 pKa = 10.57DD382 pKa = 3.78TSSSSPVSSDD392 pKa = 2.84TYY394 pKa = 10.81NVVFNTDD401 pKa = 2.96GSVQSVTDD409 pKa = 3.34STTGVTTSLSALSATIGGTSYY430 pKa = 10.41TVDD433 pKa = 3.72LSQTTLSTASSLATDD448 pKa = 4.18TTALQSDD455 pKa = 4.88SIASGTYY462 pKa = 8.82QGVEE466 pKa = 3.91IEE468 pKa = 4.4SDD470 pKa = 3.75GSVMAQFDD478 pKa = 3.92TGTQLIGKK486 pKa = 8.8IALANFANVDD496 pKa = 3.46GLNAVDD502 pKa = 3.98GQAYY506 pKa = 6.85TATASSGDD514 pKa = 3.69AKK516 pKa = 10.67IGLAGANGTGTLSVGSVEE534 pKa = 5.47SSTTDD539 pKa = 3.27LTSDD543 pKa = 3.78LSALIVAQEE552 pKa = 4.23AYY554 pKa = 10.14SANTKK559 pKa = 9.96IVTTADD565 pKa = 3.08QLLQTTIAMKK575 pKa = 10.31QQ576 pKa = 3.01
MM1 pKa = 7.41SVFNALTTAVSGINAQSTAFTNLSNNIANSQTVGYY36 pKa = 9.73KK37 pKa = 9.3ATTTAFQDD45 pKa = 4.32FVAGSLGKK53 pKa = 9.75NASSADD59 pKa = 3.27ISDD62 pKa = 3.47SVAAVTVQHH71 pKa = 6.58VDD73 pKa = 3.32NQGTAATSTDD83 pKa = 3.4SLATAISGSGLFVVSKK99 pKa = 10.61EE100 pKa = 4.29SGASTSTNTQFQSQNYY116 pKa = 4.93YY117 pKa = 8.09TRR119 pKa = 11.84NGEE122 pKa = 4.18VYY124 pKa = 10.28KK125 pKa = 11.11NNSGYY130 pKa = 10.79LVNTSGYY137 pKa = 9.17YY138 pKa = 10.07LDD140 pKa = 5.48GYY142 pKa = 9.95MVDD145 pKa = 3.85PTTGSLNASLTQLNVANVAFRR166 pKa = 11.84PTQTTTLTLSATVGTMPSSNASYY189 pKa = 8.96TAADD193 pKa = 3.57AQSYY197 pKa = 6.01TTAPVTTYY205 pKa = 10.67DD206 pKa = 4.74ASATPHH212 pKa = 6.76KK213 pKa = 10.27IALSWTQSGTNPLVWNVSAYY233 pKa = 10.49DD234 pKa = 3.66ADD236 pKa = 4.06GTGTIASNSYY246 pKa = 10.25QVTFDD251 pKa = 3.58SSGSLASVADD261 pKa = 3.63STTGKK266 pKa = 9.05TIGSTLTGASASIPITADD284 pKa = 3.28YY285 pKa = 11.22NGVAQNITLDD295 pKa = 3.52LGTIGGTSGTVMAASTGTASTNQANALTASGTTLTMSKK333 pKa = 10.43SDD335 pKa = 3.97VIGTTTGSNQTSMTSPTVANGTSVAAKK362 pKa = 6.92WTQTAASPPTWSLSLVDD379 pKa = 5.33PYY381 pKa = 10.57DD382 pKa = 3.78TSSSSPVSSDD392 pKa = 2.84TYY394 pKa = 10.81NVVFNTDD401 pKa = 2.96GSVQSVTDD409 pKa = 3.34STTGVTTSLSALSATIGGTSYY430 pKa = 10.41TVDD433 pKa = 3.72LSQTTLSTASSLATDD448 pKa = 4.18TTALQSDD455 pKa = 4.88SIASGTYY462 pKa = 8.82QGVEE466 pKa = 3.91IEE468 pKa = 4.4SDD470 pKa = 3.75GSVMAQFDD478 pKa = 3.92TGTQLIGKK486 pKa = 8.8IALANFANVDD496 pKa = 3.46GLNAVDD502 pKa = 3.98GQAYY506 pKa = 6.85TATASSGDD514 pKa = 3.69AKK516 pKa = 10.67IGLAGANGTGTLSVGSVEE534 pKa = 5.47SSTTDD539 pKa = 3.27LTSDD543 pKa = 3.78LSALIVAQEE552 pKa = 4.23AYY554 pKa = 10.14SANTKK559 pKa = 9.96IVTTADD565 pKa = 3.08QLLQTTIAMKK575 pKa = 10.31QQ576 pKa = 3.01
Molecular weight: 58.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370G1M2|A0A370G1M2_GLULI Sodium/proton antiporter (CPA1 family) OS=Gluconacetobacter liquefaciens OX=89584 GN=C7453_10940 PE=4 SV=1
MM1 pKa = 7.84DD2 pKa = 5.7RR3 pKa = 11.84DD4 pKa = 4.04GPVARR9 pKa = 11.84QGDD12 pKa = 4.01RR13 pKa = 11.84RR14 pKa = 11.84VHH16 pKa = 6.73LLLQRR21 pKa = 11.84DD22 pKa = 3.81GWAVNPKK29 pKa = 10.03RR30 pKa = 11.84IYY32 pKa = 10.41RR33 pKa = 11.84LYY35 pKa = 10.4KK36 pKa = 9.91ALGMQLRR43 pKa = 11.84NKK45 pKa = 9.35VPRR48 pKa = 11.84HH49 pKa = 4.93RR50 pKa = 11.84VTAKK54 pKa = 9.96VRR56 pKa = 11.84EE57 pKa = 4.2DD58 pKa = 3.35RR59 pKa = 11.84CPATHH64 pKa = 6.1THH66 pKa = 7.11ANDD69 pKa = 2.91SWAKK73 pKa = 10.69DD74 pKa = 3.65CVHH77 pKa = 7.18DD78 pKa = 5.46RR79 pKa = 11.84RR80 pKa = 11.84ATDD83 pKa = 3.43RR84 pKa = 11.84RR85 pKa = 11.84ILTVIDD91 pKa = 3.11IFSRR95 pKa = 11.84FSLATDD101 pKa = 3.15PRR103 pKa = 11.84FSYY106 pKa = 10.35RR107 pKa = 11.84GEE109 pKa = 3.97NVAQTLEE116 pKa = 4.52RR117 pKa = 11.84ICEE120 pKa = 3.89
MM1 pKa = 7.84DD2 pKa = 5.7RR3 pKa = 11.84DD4 pKa = 4.04GPVARR9 pKa = 11.84QGDD12 pKa = 4.01RR13 pKa = 11.84RR14 pKa = 11.84VHH16 pKa = 6.73LLLQRR21 pKa = 11.84DD22 pKa = 3.81GWAVNPKK29 pKa = 10.03RR30 pKa = 11.84IYY32 pKa = 10.41RR33 pKa = 11.84LYY35 pKa = 10.4KK36 pKa = 9.91ALGMQLRR43 pKa = 11.84NKK45 pKa = 9.35VPRR48 pKa = 11.84HH49 pKa = 4.93RR50 pKa = 11.84VTAKK54 pKa = 9.96VRR56 pKa = 11.84EE57 pKa = 4.2DD58 pKa = 3.35RR59 pKa = 11.84CPATHH64 pKa = 6.1THH66 pKa = 7.11ANDD69 pKa = 2.91SWAKK73 pKa = 10.69DD74 pKa = 3.65CVHH77 pKa = 7.18DD78 pKa = 5.46RR79 pKa = 11.84RR80 pKa = 11.84ATDD83 pKa = 3.43RR84 pKa = 11.84RR85 pKa = 11.84ILTVIDD91 pKa = 3.11IFSRR95 pKa = 11.84FSLATDD101 pKa = 3.15PRR103 pKa = 11.84FSYY106 pKa = 10.35RR107 pKa = 11.84GEE109 pKa = 3.97NVAQTLEE116 pKa = 4.52RR117 pKa = 11.84ICEE120 pKa = 3.89
Molecular weight: 14.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1239589 |
29 |
2368 |
335.4 |
36.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.724 ± 0.059 | 0.932 ± 0.015 |
5.693 ± 0.031 | 4.693 ± 0.039 |
3.443 ± 0.026 | 8.914 ± 0.04 |
2.406 ± 0.018 | 4.791 ± 0.026 |
2.126 ± 0.031 | 10.32 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.575 ± 0.018 | 2.453 ± 0.03 |
5.745 ± 0.033 | 3.253 ± 0.026 |
7.752 ± 0.044 | 5.386 ± 0.036 |
5.702 ± 0.035 | 7.378 ± 0.034 |
1.432 ± 0.016 | 2.282 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
