
Arthrobacter phage Daob
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Coralvirus; unclassified Coralvirus
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
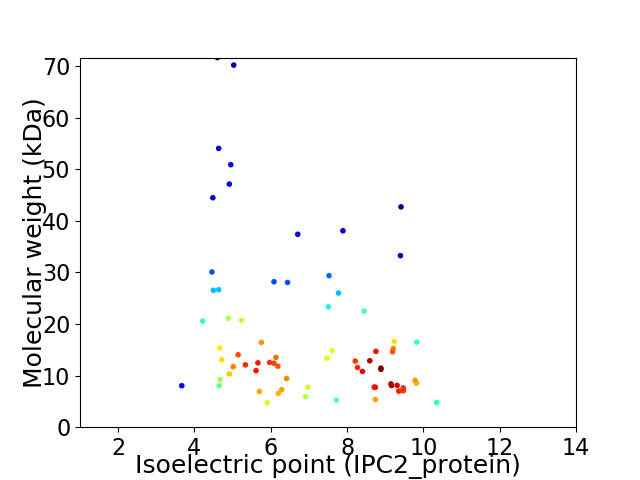
Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

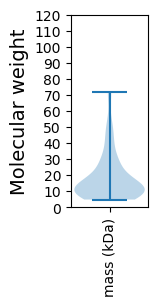
Protein with the lowest isoelectric point:
>tr|A0A3G2KFC8|A0A3G2KFC8_9CAUD Head-to-tail stopper OS=Arthrobacter phage Daob OX=2419954 GN=11 PE=4 SV=1
MM1 pKa = 7.48GNVSDD6 pKa = 4.32WEE8 pKa = 4.43VNGDD12 pKa = 3.8GFATGPVIEE21 pKa = 5.05VEE23 pKa = 4.19NGDD26 pKa = 3.71NPVTVSAVWTDD37 pKa = 3.14EE38 pKa = 5.03DD39 pKa = 4.99GVCVSVDD46 pKa = 3.62AGDD49 pKa = 4.21EE50 pKa = 3.99PLKK53 pKa = 11.13ARR55 pKa = 11.84VAYY58 pKa = 8.99RR59 pKa = 11.84VCAAIMEE66 pKa = 4.72LAAVSAPEE74 pKa = 4.29LSSSS78 pKa = 3.39
MM1 pKa = 7.48GNVSDD6 pKa = 4.32WEE8 pKa = 4.43VNGDD12 pKa = 3.8GFATGPVIEE21 pKa = 5.05VEE23 pKa = 4.19NGDD26 pKa = 3.71NPVTVSAVWTDD37 pKa = 3.14EE38 pKa = 5.03DD39 pKa = 4.99GVCVSVDD46 pKa = 3.62AGDD49 pKa = 4.21EE50 pKa = 3.99PLKK53 pKa = 11.13ARR55 pKa = 11.84VAYY58 pKa = 8.99RR59 pKa = 11.84VCAAIMEE66 pKa = 4.72LAAVSAPEE74 pKa = 4.29LSSSS78 pKa = 3.39
Molecular weight: 8.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2KFI0|A0A3G2KFI0_9CAUD MerR-like helix-turn-helix DNA binding domain protein OS=Arthrobacter phage Daob OX=2419954 GN=70 PE=4 SV=1
MM1 pKa = 7.52GRR3 pKa = 11.84PPLPVGTWGAVRR15 pKa = 11.84VEE17 pKa = 4.99RR18 pKa = 11.84IAGGYY23 pKa = 7.67RR24 pKa = 11.84ARR26 pKa = 11.84ARR28 pKa = 11.84FRR30 pKa = 11.84DD31 pKa = 3.49FDD33 pKa = 3.39GRR35 pKa = 11.84TRR37 pKa = 11.84DD38 pKa = 3.38VEE40 pKa = 4.21RR41 pKa = 11.84SGKK44 pKa = 8.31TKK46 pKa = 10.24GAARR50 pKa = 11.84AALTADD56 pKa = 3.91LNDD59 pKa = 3.45RR60 pKa = 11.84TAPAGEE66 pKa = 4.98EE67 pKa = 3.86ITASTRR73 pKa = 11.84LQLVAEE79 pKa = 4.79IWRR82 pKa = 11.84AEE84 pKa = 3.78KK85 pKa = 10.03WPNLAEE91 pKa = 4.22NSRR94 pKa = 11.84KK95 pKa = 9.81RR96 pKa = 11.84YY97 pKa = 9.47RR98 pKa = 11.84DD99 pKa = 3.27ALEE102 pKa = 4.26DD103 pKa = 4.42HH104 pKa = 6.97ILPGLGALTVSEE116 pKa = 4.59CSVTRR121 pKa = 11.84IDD123 pKa = 4.51RR124 pKa = 11.84FLKK127 pKa = 9.45ATAAGTGAPSAKK139 pKa = 9.43VCRR142 pKa = 11.84SVLSGILGLAVRR154 pKa = 11.84HH155 pKa = 5.79GAAATNPVRR164 pKa = 11.84DD165 pKa = 3.64VAGITVTPKK174 pKa = 10.38EE175 pKa = 4.0IRR177 pKa = 11.84ALTLEE182 pKa = 4.69EE183 pKa = 3.5IRR185 pKa = 11.84AARR188 pKa = 11.84SAVRR192 pKa = 11.84SWQLGEE198 pKa = 4.0PLAEE202 pKa = 4.04GRR204 pKa = 11.84PRR206 pKa = 11.84RR207 pKa = 11.84GRR209 pKa = 11.84PPTQDD214 pKa = 3.83LLDD217 pKa = 4.06ILDD220 pKa = 4.63LLLATGARR228 pKa = 11.84IGEE231 pKa = 4.23LLAIRR236 pKa = 11.84WSDD239 pKa = 3.03VDD241 pKa = 5.53LEE243 pKa = 5.04DD244 pKa = 3.69GTLTISGTIVSTEE257 pKa = 4.01DD258 pKa = 2.83KK259 pKa = 9.71PARR262 pKa = 11.84LIRR265 pKa = 11.84QAHH268 pKa = 6.06PKK270 pKa = 10.08SSTSRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84LALPPFAIDD286 pKa = 2.93ALMRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84LAVTVANVHH301 pKa = 6.45DD302 pKa = 4.25VVFPSTEE309 pKa = 3.7GTLRR313 pKa = 11.84DD314 pKa = 3.73PGSVRR319 pKa = 11.84KK320 pKa = 9.26QLAKK324 pKa = 10.57VLAPAGLGWVTPHH337 pKa = 6.16VFRR340 pKa = 11.84KK341 pKa = 7.9TVATALDD348 pKa = 3.57AAEE351 pKa = 4.78DD352 pKa = 3.77LRR354 pKa = 11.84TAADD358 pKa = 3.66QLGHH362 pKa = 7.2AGTDD366 pKa = 3.2VTRR369 pKa = 11.84RR370 pKa = 11.84HH371 pKa = 5.53YY372 pKa = 9.64VQKK375 pKa = 9.08THH377 pKa = 6.32QGPDD381 pKa = 2.84ARR383 pKa = 11.84ATLEE387 pKa = 3.9QLVRR391 pKa = 11.84PAGPP395 pKa = 3.2
MM1 pKa = 7.52GRR3 pKa = 11.84PPLPVGTWGAVRR15 pKa = 11.84VEE17 pKa = 4.99RR18 pKa = 11.84IAGGYY23 pKa = 7.67RR24 pKa = 11.84ARR26 pKa = 11.84ARR28 pKa = 11.84FRR30 pKa = 11.84DD31 pKa = 3.49FDD33 pKa = 3.39GRR35 pKa = 11.84TRR37 pKa = 11.84DD38 pKa = 3.38VEE40 pKa = 4.21RR41 pKa = 11.84SGKK44 pKa = 8.31TKK46 pKa = 10.24GAARR50 pKa = 11.84AALTADD56 pKa = 3.91LNDD59 pKa = 3.45RR60 pKa = 11.84TAPAGEE66 pKa = 4.98EE67 pKa = 3.86ITASTRR73 pKa = 11.84LQLVAEE79 pKa = 4.79IWRR82 pKa = 11.84AEE84 pKa = 3.78KK85 pKa = 10.03WPNLAEE91 pKa = 4.22NSRR94 pKa = 11.84KK95 pKa = 9.81RR96 pKa = 11.84YY97 pKa = 9.47RR98 pKa = 11.84DD99 pKa = 3.27ALEE102 pKa = 4.26DD103 pKa = 4.42HH104 pKa = 6.97ILPGLGALTVSEE116 pKa = 4.59CSVTRR121 pKa = 11.84IDD123 pKa = 4.51RR124 pKa = 11.84FLKK127 pKa = 9.45ATAAGTGAPSAKK139 pKa = 9.43VCRR142 pKa = 11.84SVLSGILGLAVRR154 pKa = 11.84HH155 pKa = 5.79GAAATNPVRR164 pKa = 11.84DD165 pKa = 3.64VAGITVTPKK174 pKa = 10.38EE175 pKa = 4.0IRR177 pKa = 11.84ALTLEE182 pKa = 4.69EE183 pKa = 3.5IRR185 pKa = 11.84AARR188 pKa = 11.84SAVRR192 pKa = 11.84SWQLGEE198 pKa = 4.0PLAEE202 pKa = 4.04GRR204 pKa = 11.84PRR206 pKa = 11.84RR207 pKa = 11.84GRR209 pKa = 11.84PPTQDD214 pKa = 3.83LLDD217 pKa = 4.06ILDD220 pKa = 4.63LLLATGARR228 pKa = 11.84IGEE231 pKa = 4.23LLAIRR236 pKa = 11.84WSDD239 pKa = 3.03VDD241 pKa = 5.53LEE243 pKa = 5.04DD244 pKa = 3.69GTLTISGTIVSTEE257 pKa = 4.01DD258 pKa = 2.83KK259 pKa = 9.71PARR262 pKa = 11.84LIRR265 pKa = 11.84QAHH268 pKa = 6.06PKK270 pKa = 10.08SSTSRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84LALPPFAIDD286 pKa = 2.93ALMRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84LAVTVANVHH301 pKa = 6.45DD302 pKa = 4.25VVFPSTEE309 pKa = 3.7GTLRR313 pKa = 11.84DD314 pKa = 3.73PGSVRR319 pKa = 11.84KK320 pKa = 9.26QLAKK324 pKa = 10.57VLAPAGLGWVTPHH337 pKa = 6.16VFRR340 pKa = 11.84KK341 pKa = 7.9TVATALDD348 pKa = 3.57AAEE351 pKa = 4.78DD352 pKa = 3.77LRR354 pKa = 11.84TAADD358 pKa = 3.66QLGHH362 pKa = 7.2AGTDD366 pKa = 3.2VTRR369 pKa = 11.84RR370 pKa = 11.84HH371 pKa = 5.53YY372 pKa = 9.64VQKK375 pKa = 9.08THH377 pKa = 6.32QGPDD381 pKa = 2.84ARR383 pKa = 11.84ATLEE387 pKa = 3.9QLVRR391 pKa = 11.84PAGPP395 pKa = 3.2
Molecular weight: 42.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12102 |
38 |
711 |
170.5 |
18.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.659 ± 0.47 | 0.835 ± 0.108 |
5.47 ± 0.172 | 6.619 ± 0.278 |
2.628 ± 0.206 | 9.808 ± 0.459 |
1.396 ± 0.138 | 3.586 ± 0.191 |
3.603 ± 0.23 | 8.982 ± 0.318 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.083 | 2.363 ± 0.152 |
5.594 ± 0.334 | 3.057 ± 0.17 |
7.784 ± 0.419 | 5.404 ± 0.242 |
5.371 ± 0.294 | 8.651 ± 0.394 |
2.115 ± 0.155 | 1.95 ± 0.149 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
