
Moorea producens 3L
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Oscillatoriaceae; Moorea; Moorea producens
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
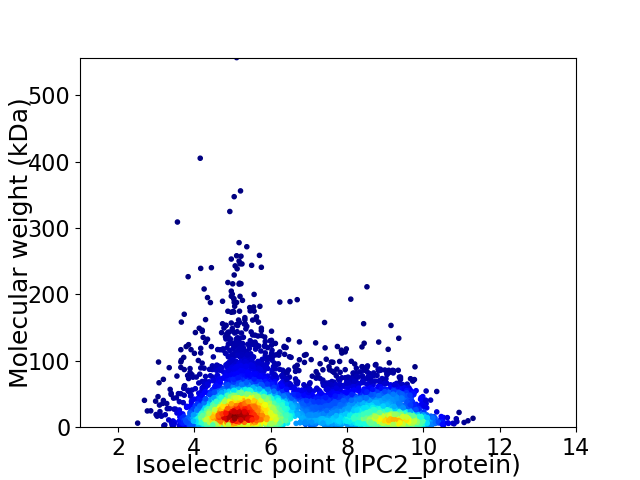
Virtual 2D-PAGE plot for 7382 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4XWZ3|F4XWZ3_9CYAN Uncharacterized protein OS=Moorea producens 3L OX=489825 GN=LYNGBM3L_46340 PE=4 SV=1
MM1 pKa = 7.72IDD3 pKa = 4.41IKK5 pKa = 9.88TSWVSYY11 pKa = 10.45QLAFLFNSFRR21 pKa = 11.84PVMKK25 pKa = 10.65SLFNYY30 pKa = 9.58LKK32 pKa = 10.38KK33 pKa = 10.43IPNHH37 pKa = 6.35SSDD40 pKa = 3.38SPRR43 pKa = 11.84ASEE46 pKa = 5.45VNSDD50 pKa = 3.39WQPLGFNLMGITNLAKK66 pKa = 9.62ATFPILVTGSALLVSTPTEE85 pKa = 3.67VAGQVGIDD93 pKa = 3.43ASLATGPRR101 pKa = 11.84PFSLRR106 pKa = 11.84YY107 pKa = 8.75SAHH110 pKa = 5.7VPGNMVAIGNASLICDD126 pKa = 3.7RR127 pKa = 11.84TNASCLNGLATGNVGNNSGGLAMQMLDD154 pKa = 3.65SDD156 pKa = 4.65SDD158 pKa = 3.75PSTFNSSSADD168 pKa = 3.19LTLPVGANVLFAGLYY183 pKa = 8.9WGGTSDD189 pKa = 4.99GATTPAPDD197 pKa = 3.18ASKK200 pKa = 10.96RR201 pKa = 11.84NEE203 pKa = 4.02ALLATPEE210 pKa = 4.23GGYY213 pKa = 7.73QTVTADD219 pKa = 3.32TFTSIDD225 pKa = 3.41NSATSGWDD233 pKa = 3.39VYY235 pKa = 11.72SSFADD240 pKa = 3.51VTSLVQAAGSGTYY253 pKa = 9.39TLANVQASTGTGFTYY268 pKa = 10.64PNAGWSLIVVYY279 pKa = 9.83EE280 pKa = 4.42DD281 pKa = 3.3PSEE284 pKa = 3.92PRR286 pKa = 11.84RR287 pKa = 11.84NMTVFDD293 pKa = 4.97GYY295 pKa = 11.3DD296 pKa = 3.16FSGFNTGNTQTLTGLRR312 pKa = 11.84TPPTPGFSVFMGAFAGDD329 pKa = 4.07GEE331 pKa = 4.83PDD333 pKa = 3.31VSGDD337 pKa = 3.39TLSINNTPVSDD348 pKa = 4.0PVNPLNNFYY357 pKa = 11.1NSTISQYY364 pKa = 10.68GSHH367 pKa = 5.23VTSRR371 pKa = 11.84TPNDD375 pKa = 4.03PYY377 pKa = 11.91NMVVDD382 pKa = 4.72IDD384 pKa = 4.92LLDD387 pKa = 3.51LTAWNEE393 pKa = 3.94ANNVIPTNATSIDD406 pKa = 4.26LNLSTSGDD414 pKa = 4.32GIWPMVYY421 pKa = 10.14FFGVEE426 pKa = 3.91VFEE429 pKa = 5.06PNLVTEE435 pKa = 4.82FEE437 pKa = 4.38KK438 pKa = 8.75TTPQTTYY445 pKa = 11.24QNGDD449 pKa = 3.65TIGYY453 pKa = 6.55TISVTNTGNDD463 pKa = 3.17NAVNTVITDD472 pKa = 4.23IIPTGTTFVPGSLKK486 pKa = 10.63INGVTKK492 pKa = 10.45TDD494 pKa = 3.36GATDD498 pKa = 3.38EE499 pKa = 5.37AEE501 pKa = 3.95FDD503 pKa = 3.57GTNVIFRR510 pKa = 11.84VGTGANTTQGGQLNVNEE527 pKa = 4.33TVTMSFEE534 pKa = 4.39VTVTAAPGEE543 pKa = 4.46KK544 pKa = 9.9VCNQASIDD552 pKa = 3.96YY553 pKa = 9.95EE554 pKa = 4.62GEE556 pKa = 3.79ASGNQASGTSDD567 pKa = 3.86DD568 pKa = 4.73PNTATFGDD576 pKa = 4.26CTEE579 pKa = 4.27ITVTPPTTTVDD590 pKa = 3.44YY591 pKa = 11.31GDD593 pKa = 5.06APDD596 pKa = 3.98TTASTGNGDD605 pKa = 4.08YY606 pKa = 8.84QTLSANSGPTHH617 pKa = 6.94TIDD620 pKa = 2.97NSTYY624 pKa = 10.41LGAGVTADD632 pKa = 3.42TDD634 pKa = 3.93GFGDD638 pKa = 4.05GTDD641 pKa = 3.7NNNNATDD648 pKa = 3.78DD649 pKa = 3.95TDD651 pKa = 5.86DD652 pKa = 3.85GVQINSASLQGQTLPISDD670 pKa = 4.2NVRR673 pKa = 11.84LDD675 pKa = 2.93ITTAGSGVLNAWIDD689 pKa = 3.48WNADD693 pKa = 2.73GDD695 pKa = 4.09FDD697 pKa = 6.37DD698 pKa = 5.72SGEE701 pKa = 4.15QIATNASPSGNAIALNLTVPAGATVGNTYY730 pKa = 11.09ARR732 pKa = 11.84FRR734 pKa = 11.84YY735 pKa = 10.26SSDD738 pKa = 2.95TGLTPTGAASDD749 pKa = 4.58GEE751 pKa = 4.38VEE753 pKa = 4.81DD754 pKa = 4.05YY755 pKa = 10.98QIAIANSTPTTASVGDD771 pKa = 4.1QVWEE775 pKa = 4.36DD776 pKa = 3.63LDD778 pKa = 5.03GDD780 pKa = 4.87GIQDD784 pKa = 3.73PGEE787 pKa = 4.15PGIPNALVNIYY798 pKa = 10.5KK799 pKa = 10.13SDD801 pKa = 3.65NTLVDD806 pKa = 3.31ATFTDD811 pKa = 3.26SSGNYY816 pKa = 9.44SFSDD820 pKa = 4.57LDD822 pKa = 3.96PGDD825 pKa = 4.27YY826 pKa = 9.96IIEE829 pKa = 4.32FTSPAVGYY837 pKa = 10.7SFTRR841 pKa = 11.84QDD843 pKa = 4.02AGDD846 pKa = 3.94DD847 pKa = 4.02TLDD850 pKa = 3.66SDD852 pKa = 4.71ADD854 pKa = 3.86PVTGRR859 pKa = 11.84TDD861 pKa = 3.38TFTLAAGDD869 pKa = 4.03NNTSIDD875 pKa = 3.43AGLTLKK881 pKa = 11.03ANTIVACDD889 pKa = 4.66IIPLQATNWTEE900 pKa = 3.81TLNVEE905 pKa = 4.61KK906 pKa = 10.27FDD908 pKa = 4.09SALGSLARR916 pKa = 11.84VDD918 pKa = 4.06LTLSTLIAQEE928 pKa = 4.62LIYY931 pKa = 10.68EE932 pKa = 4.28NTADD936 pKa = 3.65QPKK939 pKa = 9.65TITVNQEE946 pKa = 3.74GEE948 pKa = 4.47VSVSLPDD955 pKa = 3.37TNANITRR962 pKa = 11.84NYY964 pKa = 9.1NQSTDD969 pKa = 3.77FNLPAFDD976 pKa = 5.29GSDD979 pKa = 3.43PPDD982 pKa = 3.59FSGTSGGRR990 pKa = 11.84TPTILAYY997 pKa = 7.55EE998 pKa = 4.31THH1000 pKa = 5.92TEE1002 pKa = 4.19EE1003 pKa = 4.38YY1004 pKa = 10.12TNLADD1009 pKa = 5.89FISSSSPVEE1018 pKa = 4.1TVSIPVNTTSAATFINSQNASTGSSSEE1045 pKa = 3.97ATAGLCVTYY1054 pKa = 9.52TYY1056 pKa = 11.42SLVPKK1061 pKa = 9.1EE1062 pKa = 4.26PSASIVPEE1070 pKa = 3.94EE1071 pKa = 4.41ASPEE1075 pKa = 3.91AGNIVINEE1083 pKa = 3.79VLYY1086 pKa = 10.92AQTGSSAEE1094 pKa = 4.07ANDD1097 pKa = 4.05EE1098 pKa = 4.56FIEE1101 pKa = 5.29LYY1103 pKa = 10.57NSSDD1107 pKa = 3.54NAVDD1111 pKa = 3.46ISNWKK1116 pKa = 9.64LADD1119 pKa = 3.47SNLIFNSTDD1128 pKa = 2.82NTGNITGDD1136 pKa = 3.64AANPAYY1142 pKa = 9.76IFPNNTTLQPGEE1154 pKa = 4.32YY1155 pKa = 10.02AVIWIGNNTSDD1166 pKa = 4.49HH1167 pKa = 5.81QAPDD1171 pKa = 3.19AAFQDD1176 pKa = 4.13WLGKK1180 pKa = 9.48AAKK1183 pKa = 9.84LNNAGEE1189 pKa = 4.87DD1190 pKa = 2.55IWLYY1194 pKa = 11.06DD1195 pKa = 3.9EE1196 pKa = 4.43EE1197 pKa = 4.79FKK1199 pKa = 11.0IVDD1202 pKa = 3.7YY1203 pKa = 10.78IAYY1206 pKa = 9.9GSNNAINTPPSAEE1219 pKa = 4.01LNLWDD1224 pKa = 4.63DD1225 pKa = 4.34TYY1227 pKa = 11.71QRR1229 pKa = 11.84DD1230 pKa = 3.78LDD1232 pKa = 4.11GASTGQSISLTPNGEE1247 pKa = 4.32DD1248 pKa = 4.54GNTSACWEE1256 pKa = 4.23HH1257 pKa = 6.26TLSNDD1262 pKa = 3.09ANGRR1266 pKa = 11.84CPEE1269 pKa = 3.97FLEE1272 pKa = 4.56TRR1274 pKa = 11.84DD1275 pKa = 3.37TDD1277 pKa = 4.08TVSVRR1282 pKa = 11.84QTSVGVYY1289 pKa = 10.77NNILPNVILVKK1300 pKa = 10.42RR1301 pKa = 11.84ITAIKK1306 pKa = 10.17PKK1308 pKa = 9.52DD1309 pKa = 3.5TNQWIEE1315 pKa = 4.13LPQSGSPFIDD1325 pKa = 5.18GIDD1328 pKa = 3.78GGSSEE1333 pKa = 4.89NNVGSDD1339 pKa = 4.01RR1340 pKa = 11.84AADD1343 pKa = 4.0DD1344 pKa = 5.33NDD1346 pKa = 4.52PNWPNPDD1353 pKa = 2.87VYY1355 pKa = 11.19LRR1357 pKa = 11.84GLINSGQVMPGDD1369 pKa = 3.64EE1370 pKa = 4.71LEE1372 pKa = 4.23YY1373 pKa = 10.4TVYY1376 pKa = 10.58FLSNGSNDD1384 pKa = 3.3AKK1386 pKa = 10.8NLRR1389 pKa = 11.84ICDD1392 pKa = 3.85RR1393 pKa = 11.84VPGEE1397 pKa = 4.17TTFITDD1403 pKa = 3.8SFNATAGFPSSDD1415 pKa = 2.96VGIAMFQSTTALASGGPAEE1434 pKa = 4.12PNIYY1438 pKa = 8.7LTNIPDD1444 pKa = 3.62SDD1446 pKa = 3.26RR1447 pKa = 11.84GRR1449 pKa = 11.84YY1450 pKa = 8.22YY1451 pKa = 11.18GPGTSVPSGCNVTFNQNGVVVVEE1474 pKa = 4.49VGDD1477 pKa = 3.82VPNADD1482 pKa = 3.78SPGNPPNSYY1491 pKa = 10.41GFIRR1495 pKa = 11.84FRR1497 pKa = 11.84ALVKK1501 pKa = 10.72
MM1 pKa = 7.72IDD3 pKa = 4.41IKK5 pKa = 9.88TSWVSYY11 pKa = 10.45QLAFLFNSFRR21 pKa = 11.84PVMKK25 pKa = 10.65SLFNYY30 pKa = 9.58LKK32 pKa = 10.38KK33 pKa = 10.43IPNHH37 pKa = 6.35SSDD40 pKa = 3.38SPRR43 pKa = 11.84ASEE46 pKa = 5.45VNSDD50 pKa = 3.39WQPLGFNLMGITNLAKK66 pKa = 9.62ATFPILVTGSALLVSTPTEE85 pKa = 3.67VAGQVGIDD93 pKa = 3.43ASLATGPRR101 pKa = 11.84PFSLRR106 pKa = 11.84YY107 pKa = 8.75SAHH110 pKa = 5.7VPGNMVAIGNASLICDD126 pKa = 3.7RR127 pKa = 11.84TNASCLNGLATGNVGNNSGGLAMQMLDD154 pKa = 3.65SDD156 pKa = 4.65SDD158 pKa = 3.75PSTFNSSSADD168 pKa = 3.19LTLPVGANVLFAGLYY183 pKa = 8.9WGGTSDD189 pKa = 4.99GATTPAPDD197 pKa = 3.18ASKK200 pKa = 10.96RR201 pKa = 11.84NEE203 pKa = 4.02ALLATPEE210 pKa = 4.23GGYY213 pKa = 7.73QTVTADD219 pKa = 3.32TFTSIDD225 pKa = 3.41NSATSGWDD233 pKa = 3.39VYY235 pKa = 11.72SSFADD240 pKa = 3.51VTSLVQAAGSGTYY253 pKa = 9.39TLANVQASTGTGFTYY268 pKa = 10.64PNAGWSLIVVYY279 pKa = 9.83EE280 pKa = 4.42DD281 pKa = 3.3PSEE284 pKa = 3.92PRR286 pKa = 11.84RR287 pKa = 11.84NMTVFDD293 pKa = 4.97GYY295 pKa = 11.3DD296 pKa = 3.16FSGFNTGNTQTLTGLRR312 pKa = 11.84TPPTPGFSVFMGAFAGDD329 pKa = 4.07GEE331 pKa = 4.83PDD333 pKa = 3.31VSGDD337 pKa = 3.39TLSINNTPVSDD348 pKa = 4.0PVNPLNNFYY357 pKa = 11.1NSTISQYY364 pKa = 10.68GSHH367 pKa = 5.23VTSRR371 pKa = 11.84TPNDD375 pKa = 4.03PYY377 pKa = 11.91NMVVDD382 pKa = 4.72IDD384 pKa = 4.92LLDD387 pKa = 3.51LTAWNEE393 pKa = 3.94ANNVIPTNATSIDD406 pKa = 4.26LNLSTSGDD414 pKa = 4.32GIWPMVYY421 pKa = 10.14FFGVEE426 pKa = 3.91VFEE429 pKa = 5.06PNLVTEE435 pKa = 4.82FEE437 pKa = 4.38KK438 pKa = 8.75TTPQTTYY445 pKa = 11.24QNGDD449 pKa = 3.65TIGYY453 pKa = 6.55TISVTNTGNDD463 pKa = 3.17NAVNTVITDD472 pKa = 4.23IIPTGTTFVPGSLKK486 pKa = 10.63INGVTKK492 pKa = 10.45TDD494 pKa = 3.36GATDD498 pKa = 3.38EE499 pKa = 5.37AEE501 pKa = 3.95FDD503 pKa = 3.57GTNVIFRR510 pKa = 11.84VGTGANTTQGGQLNVNEE527 pKa = 4.33TVTMSFEE534 pKa = 4.39VTVTAAPGEE543 pKa = 4.46KK544 pKa = 9.9VCNQASIDD552 pKa = 3.96YY553 pKa = 9.95EE554 pKa = 4.62GEE556 pKa = 3.79ASGNQASGTSDD567 pKa = 3.86DD568 pKa = 4.73PNTATFGDD576 pKa = 4.26CTEE579 pKa = 4.27ITVTPPTTTVDD590 pKa = 3.44YY591 pKa = 11.31GDD593 pKa = 5.06APDD596 pKa = 3.98TTASTGNGDD605 pKa = 4.08YY606 pKa = 8.84QTLSANSGPTHH617 pKa = 6.94TIDD620 pKa = 2.97NSTYY624 pKa = 10.41LGAGVTADD632 pKa = 3.42TDD634 pKa = 3.93GFGDD638 pKa = 4.05GTDD641 pKa = 3.7NNNNATDD648 pKa = 3.78DD649 pKa = 3.95TDD651 pKa = 5.86DD652 pKa = 3.85GVQINSASLQGQTLPISDD670 pKa = 4.2NVRR673 pKa = 11.84LDD675 pKa = 2.93ITTAGSGVLNAWIDD689 pKa = 3.48WNADD693 pKa = 2.73GDD695 pKa = 4.09FDD697 pKa = 6.37DD698 pKa = 5.72SGEE701 pKa = 4.15QIATNASPSGNAIALNLTVPAGATVGNTYY730 pKa = 11.09ARR732 pKa = 11.84FRR734 pKa = 11.84YY735 pKa = 10.26SSDD738 pKa = 2.95TGLTPTGAASDD749 pKa = 4.58GEE751 pKa = 4.38VEE753 pKa = 4.81DD754 pKa = 4.05YY755 pKa = 10.98QIAIANSTPTTASVGDD771 pKa = 4.1QVWEE775 pKa = 4.36DD776 pKa = 3.63LDD778 pKa = 5.03GDD780 pKa = 4.87GIQDD784 pKa = 3.73PGEE787 pKa = 4.15PGIPNALVNIYY798 pKa = 10.5KK799 pKa = 10.13SDD801 pKa = 3.65NTLVDD806 pKa = 3.31ATFTDD811 pKa = 3.26SSGNYY816 pKa = 9.44SFSDD820 pKa = 4.57LDD822 pKa = 3.96PGDD825 pKa = 4.27YY826 pKa = 9.96IIEE829 pKa = 4.32FTSPAVGYY837 pKa = 10.7SFTRR841 pKa = 11.84QDD843 pKa = 4.02AGDD846 pKa = 3.94DD847 pKa = 4.02TLDD850 pKa = 3.66SDD852 pKa = 4.71ADD854 pKa = 3.86PVTGRR859 pKa = 11.84TDD861 pKa = 3.38TFTLAAGDD869 pKa = 4.03NNTSIDD875 pKa = 3.43AGLTLKK881 pKa = 11.03ANTIVACDD889 pKa = 4.66IIPLQATNWTEE900 pKa = 3.81TLNVEE905 pKa = 4.61KK906 pKa = 10.27FDD908 pKa = 4.09SALGSLARR916 pKa = 11.84VDD918 pKa = 4.06LTLSTLIAQEE928 pKa = 4.62LIYY931 pKa = 10.68EE932 pKa = 4.28NTADD936 pKa = 3.65QPKK939 pKa = 9.65TITVNQEE946 pKa = 3.74GEE948 pKa = 4.47VSVSLPDD955 pKa = 3.37TNANITRR962 pKa = 11.84NYY964 pKa = 9.1NQSTDD969 pKa = 3.77FNLPAFDD976 pKa = 5.29GSDD979 pKa = 3.43PPDD982 pKa = 3.59FSGTSGGRR990 pKa = 11.84TPTILAYY997 pKa = 7.55EE998 pKa = 4.31THH1000 pKa = 5.92TEE1002 pKa = 4.19EE1003 pKa = 4.38YY1004 pKa = 10.12TNLADD1009 pKa = 5.89FISSSSPVEE1018 pKa = 4.1TVSIPVNTTSAATFINSQNASTGSSSEE1045 pKa = 3.97ATAGLCVTYY1054 pKa = 9.52TYY1056 pKa = 11.42SLVPKK1061 pKa = 9.1EE1062 pKa = 4.26PSASIVPEE1070 pKa = 3.94EE1071 pKa = 4.41ASPEE1075 pKa = 3.91AGNIVINEE1083 pKa = 3.79VLYY1086 pKa = 10.92AQTGSSAEE1094 pKa = 4.07ANDD1097 pKa = 4.05EE1098 pKa = 4.56FIEE1101 pKa = 5.29LYY1103 pKa = 10.57NSSDD1107 pKa = 3.54NAVDD1111 pKa = 3.46ISNWKK1116 pKa = 9.64LADD1119 pKa = 3.47SNLIFNSTDD1128 pKa = 2.82NTGNITGDD1136 pKa = 3.64AANPAYY1142 pKa = 9.76IFPNNTTLQPGEE1154 pKa = 4.32YY1155 pKa = 10.02AVIWIGNNTSDD1166 pKa = 4.49HH1167 pKa = 5.81QAPDD1171 pKa = 3.19AAFQDD1176 pKa = 4.13WLGKK1180 pKa = 9.48AAKK1183 pKa = 9.84LNNAGEE1189 pKa = 4.87DD1190 pKa = 2.55IWLYY1194 pKa = 11.06DD1195 pKa = 3.9EE1196 pKa = 4.43EE1197 pKa = 4.79FKK1199 pKa = 11.0IVDD1202 pKa = 3.7YY1203 pKa = 10.78IAYY1206 pKa = 9.9GSNNAINTPPSAEE1219 pKa = 4.01LNLWDD1224 pKa = 4.63DD1225 pKa = 4.34TYY1227 pKa = 11.71QRR1229 pKa = 11.84DD1230 pKa = 3.78LDD1232 pKa = 4.11GASTGQSISLTPNGEE1247 pKa = 4.32DD1248 pKa = 4.54GNTSACWEE1256 pKa = 4.23HH1257 pKa = 6.26TLSNDD1262 pKa = 3.09ANGRR1266 pKa = 11.84CPEE1269 pKa = 3.97FLEE1272 pKa = 4.56TRR1274 pKa = 11.84DD1275 pKa = 3.37TDD1277 pKa = 4.08TVSVRR1282 pKa = 11.84QTSVGVYY1289 pKa = 10.77NNILPNVILVKK1300 pKa = 10.42RR1301 pKa = 11.84ITAIKK1306 pKa = 10.17PKK1308 pKa = 9.52DD1309 pKa = 3.5TNQWIEE1315 pKa = 4.13LPQSGSPFIDD1325 pKa = 5.18GIDD1328 pKa = 3.78GGSSEE1333 pKa = 4.89NNVGSDD1339 pKa = 4.01RR1340 pKa = 11.84AADD1343 pKa = 4.0DD1344 pKa = 5.33NDD1346 pKa = 4.52PNWPNPDD1353 pKa = 2.87VYY1355 pKa = 11.19LRR1357 pKa = 11.84GLINSGQVMPGDD1369 pKa = 3.64EE1370 pKa = 4.71LEE1372 pKa = 4.23YY1373 pKa = 10.4TVYY1376 pKa = 10.58FLSNGSNDD1384 pKa = 3.3AKK1386 pKa = 10.8NLRR1389 pKa = 11.84ICDD1392 pKa = 3.85RR1393 pKa = 11.84VPGEE1397 pKa = 4.17TTFITDD1403 pKa = 3.8SFNATAGFPSSDD1415 pKa = 2.96VGIAMFQSTTALASGGPAEE1434 pKa = 4.12PNIYY1438 pKa = 8.7LTNIPDD1444 pKa = 3.62SDD1446 pKa = 3.26RR1447 pKa = 11.84GRR1449 pKa = 11.84YY1450 pKa = 8.22YY1451 pKa = 11.18GPGTSVPSGCNVTFNQNGVVVVEE1474 pKa = 4.49VGDD1477 pKa = 3.82VPNADD1482 pKa = 3.78SPGNPPNSYY1491 pKa = 10.41GFIRR1495 pKa = 11.84FRR1497 pKa = 11.84ALVKK1501 pKa = 10.72
Molecular weight: 158.39 kDa
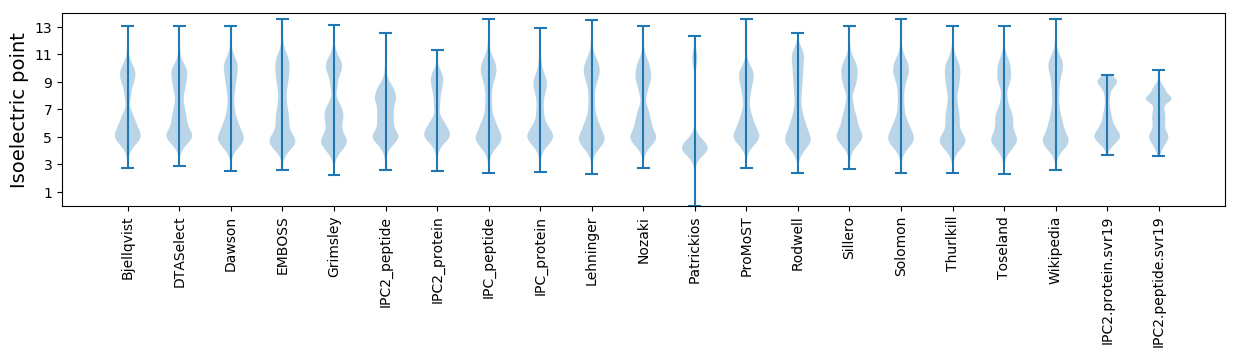
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4XIZ2|F4XIZ2_9CYAN Uncharacterized protein OS=Moorea producens 3L OX=489825 GN=LYNGBM3L_04520 PE=4 SV=1
MM1 pKa = 7.84SIQQSAVSGQRR12 pKa = 11.84SAVSGQHH19 pKa = 5.67SAVSGQRR26 pKa = 11.84SAFSGQRR33 pKa = 11.84SAVSIQRR40 pKa = 11.84SAVSGQHH47 pKa = 5.67SAVSGQRR54 pKa = 11.84SAFSGQRR61 pKa = 11.84SAVSIQRR68 pKa = 11.84SAVSGQHH75 pKa = 5.67SAVSGQRR82 pKa = 11.84SAFSGQRR89 pKa = 11.84SAVSIQRR96 pKa = 11.84SAVSGQHH103 pKa = 5.67SAVSGQRR110 pKa = 11.84SAFSGQRR117 pKa = 11.84SAVSIQRR124 pKa = 11.84SAVSGQQTT132 pKa = 3.04
MM1 pKa = 7.84SIQQSAVSGQRR12 pKa = 11.84SAVSGQHH19 pKa = 5.67SAVSGQRR26 pKa = 11.84SAFSGQRR33 pKa = 11.84SAVSIQRR40 pKa = 11.84SAVSGQHH47 pKa = 5.67SAVSGQRR54 pKa = 11.84SAFSGQRR61 pKa = 11.84SAVSIQRR68 pKa = 11.84SAVSGQHH75 pKa = 5.67SAVSGQRR82 pKa = 11.84SAFSGQRR89 pKa = 11.84SAVSIQRR96 pKa = 11.84SAVSGQHH103 pKa = 5.67SAVSGQRR110 pKa = 11.84SAFSGQRR117 pKa = 11.84SAVSIQRR124 pKa = 11.84SAVSGQQTT132 pKa = 3.04
Molecular weight: 13.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2118520 |
15 |
4862 |
287.0 |
32.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.434 ± 0.028 | 1.038 ± 0.012 |
5.111 ± 0.032 | 6.201 ± 0.03 |
3.823 ± 0.022 | 6.801 ± 0.035 |
1.861 ± 0.014 | 6.497 ± 0.028 |
5.208 ± 0.034 | 10.921 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.837 ± 0.015 | 4.4 ± 0.027 |
4.818 ± 0.025 | 5.437 ± 0.031 |
5.123 ± 0.026 | 6.675 ± 0.024 |
5.588 ± 0.027 | 6.491 ± 0.024 |
1.515 ± 0.013 | 3.127 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
