
Rubidibacter lacunae KORDI 51-2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Aphanothecaceae; Rubidibacter; Rubidibacter lacunae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
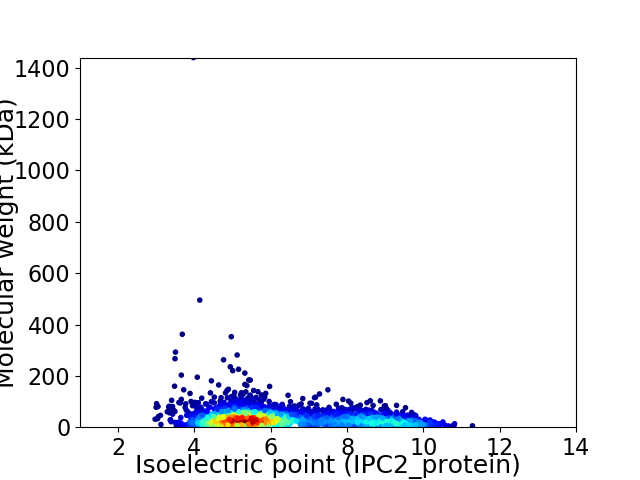
Virtual 2D-PAGE plot for 3455 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5DC01|U5DC01_9CHRO Malate dehydrogenase OS=Rubidibacter lacunae KORDI 51-2 OX=582515 GN=mdh PE=3 SV=1
MM1 pKa = 7.45IANHH5 pKa = 5.74VKK7 pKa = 10.62ALLHH11 pKa = 6.97KK12 pKa = 10.53NGCHH16 pKa = 6.11WNTGGSVGNSSGFGGSAAITYY37 pKa = 10.47SFMDD41 pKa = 3.72GRR43 pKa = 11.84PAGFDD48 pKa = 3.22PDD50 pKa = 3.7QFQDD54 pKa = 3.59FADD57 pKa = 5.93FSATQASWIEE67 pKa = 3.99HH68 pKa = 6.28AMDD71 pKa = 3.97HH72 pKa = 6.15YY73 pKa = 11.61EE74 pKa = 4.41EE75 pKa = 4.47IANITFAEE83 pKa = 4.56VASGGDD89 pKa = 3.33IDD91 pKa = 3.79WGYY94 pKa = 11.33AQTFYY99 pKa = 11.31DD100 pKa = 3.51WDD102 pKa = 3.86GDD104 pKa = 4.16GVIEE108 pKa = 4.15SHH110 pKa = 6.02EE111 pKa = 4.32VAGGLATRR119 pKa = 11.84PYY121 pKa = 10.16SNGSIIVIDD130 pKa = 3.67RR131 pKa = 11.84DD132 pKa = 3.72STNFSQGSWGYY143 pKa = 10.17LVLMHH148 pKa = 6.61EE149 pKa = 5.26LGHH152 pKa = 6.67AAAGFNDD159 pKa = 3.55VTIGLNFSEE168 pKa = 5.46TYY170 pKa = 7.19TTSQEE175 pKa = 3.89YY176 pKa = 9.27WNQGIDD182 pKa = 3.77GQVLSVSEE190 pKa = 4.52DD191 pKa = 3.26SRR193 pKa = 11.84KK194 pKa = 7.6YY195 pKa = 9.27TAMSYY200 pKa = 11.03NNHH203 pKa = 6.66PDD205 pKa = 3.12MPGVHH210 pKa = 7.16PNTLLLYY217 pKa = 10.45DD218 pKa = 4.02IAALQTLYY226 pKa = 11.07GANTSTRR233 pKa = 11.84AGNTTYY239 pKa = 10.73SWATNEE245 pKa = 4.37TFLEE249 pKa = 4.77TIWDD253 pKa = 3.82GGGTDD258 pKa = 4.51TISASNQTRR267 pKa = 11.84DD268 pKa = 3.52AVINLNPGTFSSIGSYY284 pKa = 10.31AGRR287 pKa = 11.84DD288 pKa = 3.48ATDD291 pKa = 3.23NLAIAFNVTIEE302 pKa = 3.96NAYY305 pKa = 10.04GGSGNDD311 pKa = 2.67RR312 pKa = 11.84VFGNAVGNEE321 pKa = 3.66IRR323 pKa = 11.84GGAGNDD329 pKa = 2.99RR330 pKa = 11.84LFGQDD335 pKa = 2.88GNDD338 pKa = 3.08RR339 pKa = 11.84LFGEE343 pKa = 5.12DD344 pKa = 3.3GNDD347 pKa = 3.06RR348 pKa = 11.84LFGQDD353 pKa = 4.2GNDD356 pKa = 3.4FLYY359 pKa = 10.72GQDD362 pKa = 4.25GNDD365 pKa = 3.41FLYY368 pKa = 10.73GQDD371 pKa = 4.31GDD373 pKa = 4.2DD374 pKa = 3.84FLYY377 pKa = 10.78GHH379 pKa = 7.95DD380 pKa = 3.97DD381 pKa = 3.4TDD383 pKa = 3.84RR384 pKa = 11.84LSGEE388 pKa = 4.44DD389 pKa = 3.86GNDD392 pKa = 2.87RR393 pKa = 11.84LYY395 pKa = 11.14GQEE398 pKa = 4.55GNDD401 pKa = 3.51FLYY404 pKa = 10.88GGNGDD409 pKa = 4.07DD410 pKa = 4.57FLYY413 pKa = 10.56GQEE416 pKa = 4.73GNDD419 pKa = 3.51FLYY422 pKa = 10.88GGNGDD427 pKa = 4.27DD428 pKa = 3.89RR429 pKa = 11.84QSGGKK434 pKa = 10.21GDD436 pKa = 3.75DD437 pKa = 3.13RR438 pKa = 11.84LYY440 pKa = 11.43GEE442 pKa = 4.83NGNDD446 pKa = 3.37FLYY449 pKa = 11.17GEE451 pKa = 4.76NGNDD455 pKa = 3.11SLYY458 pKa = 11.04GGKK461 pKa = 10.39GDD463 pKa = 4.33DD464 pKa = 3.48RR465 pKa = 11.84LYY467 pKa = 11.41GKK469 pKa = 10.07DD470 pKa = 4.14GNDD473 pKa = 4.45SLYY476 pKa = 10.81GWQGNDD482 pKa = 2.75RR483 pKa = 11.84LYY485 pKa = 11.43GEE487 pKa = 4.97NGNDD491 pKa = 3.19SLYY494 pKa = 11.31GEE496 pKa = 4.88NGNDD500 pKa = 3.11SLYY503 pKa = 10.85GGKK506 pKa = 10.62GSDD509 pKa = 3.62VLIGGAGADD518 pKa = 3.54TLVGFGSSSSQFDD531 pKa = 3.92TLTGGAGTDD540 pKa = 3.51TFVLGDD546 pKa = 4.29LGSVFYY552 pKa = 10.74LGSGHH557 pKa = 7.22ARR559 pKa = 11.84ITDD562 pKa = 4.07FTSGSLAIEE571 pKa = 3.94DD572 pKa = 4.42TIQIRR577 pKa = 11.84GVLNDD582 pKa = 3.29YY583 pKa = 8.41TLNQSVNYY591 pKa = 9.7GGAAALDD598 pKa = 3.7TAIFLGSDD606 pKa = 4.23LIGVVEE612 pKa = 4.65DD613 pKa = 3.77TTAIALTADD622 pKa = 3.51YY623 pKa = 7.31FTTVV627 pKa = 2.69
MM1 pKa = 7.45IANHH5 pKa = 5.74VKK7 pKa = 10.62ALLHH11 pKa = 6.97KK12 pKa = 10.53NGCHH16 pKa = 6.11WNTGGSVGNSSGFGGSAAITYY37 pKa = 10.47SFMDD41 pKa = 3.72GRR43 pKa = 11.84PAGFDD48 pKa = 3.22PDD50 pKa = 3.7QFQDD54 pKa = 3.59FADD57 pKa = 5.93FSATQASWIEE67 pKa = 3.99HH68 pKa = 6.28AMDD71 pKa = 3.97HH72 pKa = 6.15YY73 pKa = 11.61EE74 pKa = 4.41EE75 pKa = 4.47IANITFAEE83 pKa = 4.56VASGGDD89 pKa = 3.33IDD91 pKa = 3.79WGYY94 pKa = 11.33AQTFYY99 pKa = 11.31DD100 pKa = 3.51WDD102 pKa = 3.86GDD104 pKa = 4.16GVIEE108 pKa = 4.15SHH110 pKa = 6.02EE111 pKa = 4.32VAGGLATRR119 pKa = 11.84PYY121 pKa = 10.16SNGSIIVIDD130 pKa = 3.67RR131 pKa = 11.84DD132 pKa = 3.72STNFSQGSWGYY143 pKa = 10.17LVLMHH148 pKa = 6.61EE149 pKa = 5.26LGHH152 pKa = 6.67AAAGFNDD159 pKa = 3.55VTIGLNFSEE168 pKa = 5.46TYY170 pKa = 7.19TTSQEE175 pKa = 3.89YY176 pKa = 9.27WNQGIDD182 pKa = 3.77GQVLSVSEE190 pKa = 4.52DD191 pKa = 3.26SRR193 pKa = 11.84KK194 pKa = 7.6YY195 pKa = 9.27TAMSYY200 pKa = 11.03NNHH203 pKa = 6.66PDD205 pKa = 3.12MPGVHH210 pKa = 7.16PNTLLLYY217 pKa = 10.45DD218 pKa = 4.02IAALQTLYY226 pKa = 11.07GANTSTRR233 pKa = 11.84AGNTTYY239 pKa = 10.73SWATNEE245 pKa = 4.37TFLEE249 pKa = 4.77TIWDD253 pKa = 3.82GGGTDD258 pKa = 4.51TISASNQTRR267 pKa = 11.84DD268 pKa = 3.52AVINLNPGTFSSIGSYY284 pKa = 10.31AGRR287 pKa = 11.84DD288 pKa = 3.48ATDD291 pKa = 3.23NLAIAFNVTIEE302 pKa = 3.96NAYY305 pKa = 10.04GGSGNDD311 pKa = 2.67RR312 pKa = 11.84VFGNAVGNEE321 pKa = 3.66IRR323 pKa = 11.84GGAGNDD329 pKa = 2.99RR330 pKa = 11.84LFGQDD335 pKa = 2.88GNDD338 pKa = 3.08RR339 pKa = 11.84LFGEE343 pKa = 5.12DD344 pKa = 3.3GNDD347 pKa = 3.06RR348 pKa = 11.84LFGQDD353 pKa = 4.2GNDD356 pKa = 3.4FLYY359 pKa = 10.72GQDD362 pKa = 4.25GNDD365 pKa = 3.41FLYY368 pKa = 10.73GQDD371 pKa = 4.31GDD373 pKa = 4.2DD374 pKa = 3.84FLYY377 pKa = 10.78GHH379 pKa = 7.95DD380 pKa = 3.97DD381 pKa = 3.4TDD383 pKa = 3.84RR384 pKa = 11.84LSGEE388 pKa = 4.44DD389 pKa = 3.86GNDD392 pKa = 2.87RR393 pKa = 11.84LYY395 pKa = 11.14GQEE398 pKa = 4.55GNDD401 pKa = 3.51FLYY404 pKa = 10.88GGNGDD409 pKa = 4.07DD410 pKa = 4.57FLYY413 pKa = 10.56GQEE416 pKa = 4.73GNDD419 pKa = 3.51FLYY422 pKa = 10.88GGNGDD427 pKa = 4.27DD428 pKa = 3.89RR429 pKa = 11.84QSGGKK434 pKa = 10.21GDD436 pKa = 3.75DD437 pKa = 3.13RR438 pKa = 11.84LYY440 pKa = 11.43GEE442 pKa = 4.83NGNDD446 pKa = 3.37FLYY449 pKa = 11.17GEE451 pKa = 4.76NGNDD455 pKa = 3.11SLYY458 pKa = 11.04GGKK461 pKa = 10.39GDD463 pKa = 4.33DD464 pKa = 3.48RR465 pKa = 11.84LYY467 pKa = 11.41GKK469 pKa = 10.07DD470 pKa = 4.14GNDD473 pKa = 4.45SLYY476 pKa = 10.81GWQGNDD482 pKa = 2.75RR483 pKa = 11.84LYY485 pKa = 11.43GEE487 pKa = 4.97NGNDD491 pKa = 3.19SLYY494 pKa = 11.31GEE496 pKa = 4.88NGNDD500 pKa = 3.11SLYY503 pKa = 10.85GGKK506 pKa = 10.62GSDD509 pKa = 3.62VLIGGAGADD518 pKa = 3.54TLVGFGSSSSQFDD531 pKa = 3.92TLTGGAGTDD540 pKa = 3.51TFVLGDD546 pKa = 4.29LGSVFYY552 pKa = 10.74LGSGHH557 pKa = 7.22ARR559 pKa = 11.84ITDD562 pKa = 4.07FTSGSLAIEE571 pKa = 3.94DD572 pKa = 4.42TIQIRR577 pKa = 11.84GVLNDD582 pKa = 3.29YY583 pKa = 8.41TLNQSVNYY591 pKa = 9.7GGAAALDD598 pKa = 3.7TAIFLGSDD606 pKa = 4.23LIGVVEE612 pKa = 4.65DD613 pKa = 3.77TTAIALTADD622 pKa = 3.51YY623 pKa = 7.31FTTVV627 pKa = 2.69
Molecular weight: 66.83 kDa
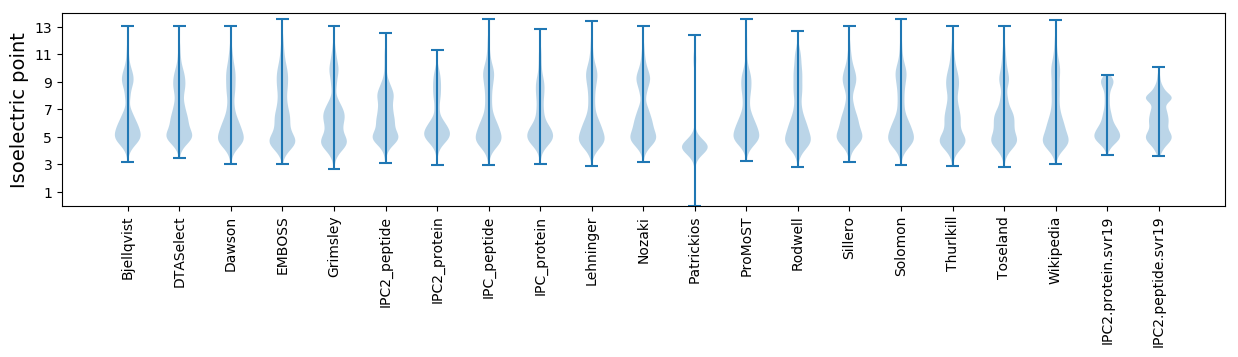
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5DNG3|U5DNG3_9CHRO Uncharacterized protein OS=Rubidibacter lacunae KORDI 51-2 OX=582515 GN=KR51_00009370 PE=4 SV=1
MM1 pKa = 7.37AQQTLRR7 pKa = 11.84GTRR10 pKa = 11.84LKK12 pKa = 10.5QKK14 pKa = 10.2RR15 pKa = 11.84KK16 pKa = 9.06LGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 9.65TGRR29 pKa = 11.84RR30 pKa = 11.84VINARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.42GRR40 pKa = 11.84ARR42 pKa = 11.84LSVV45 pKa = 3.12
MM1 pKa = 7.37AQQTLRR7 pKa = 11.84GTRR10 pKa = 11.84LKK12 pKa = 10.5QKK14 pKa = 10.2RR15 pKa = 11.84KK16 pKa = 9.06LGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 9.65TGRR29 pKa = 11.84RR30 pKa = 11.84VINARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.42GRR40 pKa = 11.84ARR42 pKa = 11.84LSVV45 pKa = 3.12
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1058787 |
24 |
13395 |
306.5 |
33.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.756 ± 0.061 | 1.138 ± 0.022 |
5.787 ± 0.062 | 5.975 ± 0.04 |
3.763 ± 0.026 | 7.658 ± 0.055 |
1.834 ± 0.028 | 5.175 ± 0.032 |
2.737 ± 0.041 | 11.224 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.684 ± 0.025 | 2.987 ± 0.034 |
5.315 ± 0.035 | 4.187 ± 0.039 |
7.079 ± 0.053 | 5.993 ± 0.033 |
5.419 ± 0.055 | 7.153 ± 0.04 |
1.488 ± 0.021 | 2.648 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
