
Streptomyces sp. BK161
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
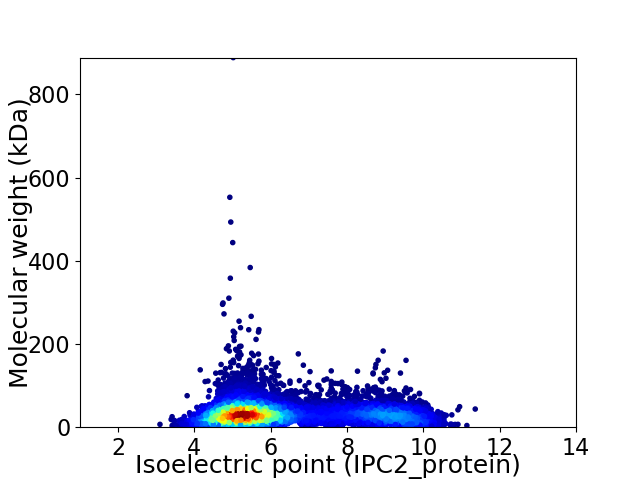
Virtual 2D-PAGE plot for 9221 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V3EKB4|A0A4V3EKB4_9ACTN Putative membrane protein YphA (DoxX/SURF4 family) OS=Streptomyces sp. BK161 OX=887463 GN=EV573_105583 PE=4 SV=1
MM1 pKa = 7.81TDD3 pKa = 3.42DD4 pKa = 3.32QHH6 pKa = 9.59GEE8 pKa = 4.05GTPQDD13 pKa = 3.66GGRR16 pKa = 11.84WDD18 pKa = 4.56PLPQGDD24 pKa = 4.33YY25 pKa = 11.28DD26 pKa = 4.37DD27 pKa = 4.9GATAFVQLPEE37 pKa = 4.38GGIDD41 pKa = 3.5ALLSGDD47 pKa = 4.46SPLAAPGHH55 pKa = 6.43GYY57 pKa = 10.04VPPQITATPPAHH69 pKa = 7.47DD70 pKa = 4.26GTDD73 pKa = 3.5PAAGAWGAPAGGTGWPDD90 pKa = 3.52PNAVPAQEE98 pKa = 4.6PGAGDD103 pKa = 3.56RR104 pKa = 11.84FTYY107 pKa = 10.44HH108 pKa = 7.21PGATGQWNFGDD119 pKa = 3.92ANGPEE124 pKa = 4.39PAPGPGHH131 pKa = 7.3DD132 pKa = 4.16VTGQWSIPVAGGDD145 pKa = 3.84LPDD148 pKa = 3.73EE149 pKa = 4.58SGEE152 pKa = 4.18YY153 pKa = 8.72STSALLEE160 pKa = 4.13QWGGTPPATLPGGAPAPWATGPGGAGGEE188 pKa = 4.27PWGQTAPSADD198 pKa = 4.65PYY200 pKa = 10.53AQPPGPYY207 pKa = 9.88DD208 pKa = 3.2SGAPGLPGGDD218 pKa = 3.0AHH220 pKa = 7.24GGPSPAAGAAEE231 pKa = 4.1AEE233 pKa = 4.67RR234 pKa = 11.84YY235 pKa = 9.79ASAADD240 pKa = 3.47AALDD244 pKa = 3.93EE245 pKa = 4.44AHH247 pKa = 6.34TAAEE251 pKa = 4.31ASSTAAVYY259 pKa = 7.47EE260 pKa = 4.41TFTGSPALPGATDD273 pKa = 3.16AHH275 pKa = 6.76AGHH278 pKa = 6.77GPGYY282 pKa = 10.35GRR284 pKa = 11.84QSEE287 pKa = 4.3QTFGQGVEE295 pKa = 4.51FTFGEE300 pKa = 4.53SGAEE304 pKa = 3.81QPPRR308 pKa = 11.84ASGAGTAYY316 pKa = 8.9DD317 pKa = 3.88TAPGSAYY324 pKa = 10.19DD325 pKa = 4.04SPSGLPEE332 pKa = 3.97EE333 pKa = 4.85ASSGFPDD340 pKa = 3.88APTDD344 pKa = 3.88GFPDD348 pKa = 4.05TPASSTEE355 pKa = 3.75QTEE358 pKa = 4.35AQQEE362 pKa = 4.54EE363 pKa = 5.19PGTPHH368 pKa = 6.85GSVSGVADD376 pKa = 4.02ATPSATDD383 pKa = 3.72GSTSGPEE390 pKa = 3.65GDD392 pKa = 4.81APPGFPGEE400 pKa = 4.31SALGAEE406 pKa = 5.06GPEE409 pKa = 4.03EE410 pKa = 4.24GAPSGAADD418 pKa = 4.32GSDD421 pKa = 3.28SGADD425 pKa = 3.22AAVTADD431 pKa = 4.13FPDD434 pKa = 3.67GRR436 pKa = 11.84APGDD440 pKa = 3.38GDD442 pKa = 3.77GTPEE446 pKa = 4.27GLPDD450 pKa = 4.11GAASDD455 pKa = 4.47ASPPLPPEE463 pKa = 4.2RR464 pKa = 11.84PAAAEE469 pKa = 3.67DD470 pKa = 3.47TGANEE475 pKa = 4.27EE476 pKa = 4.32SPEE479 pKa = 3.99YY480 pKa = 10.03PEE482 pKa = 4.86SRR484 pKa = 11.84EE485 pKa = 4.12SQASSASPDD494 pKa = 3.64GVPDD498 pKa = 3.45ATDD501 pKa = 3.22VVDD504 pKa = 5.28ASDD507 pKa = 4.28GADD510 pKa = 3.49PAPGDD515 pKa = 3.78GADD518 pKa = 4.56PSPEE522 pKa = 4.04PAPAAVPDD530 pKa = 3.74AATPHH535 pKa = 7.06DD536 pKa = 4.77APAPPQEE543 pKa = 4.05EE544 pKa = 4.67HH545 pKa = 6.88PLASYY550 pKa = 9.16VLRR553 pKa = 11.84VNGVDD558 pKa = 4.84RR559 pKa = 11.84PVTDD563 pKa = 2.96AWIGEE568 pKa = 4.15SLLYY572 pKa = 10.18VLRR575 pKa = 11.84EE576 pKa = 3.94RR577 pKa = 11.84LGLAGAKK584 pKa = 9.72DD585 pKa = 3.75GCSQGEE591 pKa = 4.12CGACNVQVDD600 pKa = 3.94GRR602 pKa = 11.84LVASCLVPSVTAAGSEE618 pKa = 4.0VRR620 pKa = 11.84TVEE623 pKa = 3.85GLAVDD628 pKa = 4.6GRR630 pKa = 11.84PSDD633 pKa = 3.66VQRR636 pKa = 11.84ALARR640 pKa = 11.84CGAVQCGFCVPGMAMTVHH658 pKa = 7.27DD659 pKa = 5.16LLEE662 pKa = 4.78GNPAPTEE669 pKa = 4.08LEE671 pKa = 4.08TRR673 pKa = 11.84QALCGNLCRR682 pKa = 11.84CSGYY686 pKa = 10.36RR687 pKa = 11.84GVLEE691 pKa = 3.9AVKK694 pKa = 10.18EE695 pKa = 4.37VVAEE699 pKa = 4.43RR700 pKa = 11.84EE701 pKa = 3.97AHH703 pKa = 5.13TAADD707 pKa = 4.16GEE709 pKa = 4.63TDD711 pKa = 3.16TGEE714 pKa = 4.3ARR716 pKa = 11.84IPHH719 pKa = 5.39QAGPGAGGVHH729 pKa = 6.81PSAFDD734 pKa = 3.46SPGFPSANGPSGPQGHH750 pKa = 6.63TDD752 pKa = 2.52SSGPHH757 pKa = 6.08DD758 pKa = 3.55QAYY761 pKa = 9.56GQDD764 pKa = 3.54GGQAA768 pKa = 3.33
MM1 pKa = 7.81TDD3 pKa = 3.42DD4 pKa = 3.32QHH6 pKa = 9.59GEE8 pKa = 4.05GTPQDD13 pKa = 3.66GGRR16 pKa = 11.84WDD18 pKa = 4.56PLPQGDD24 pKa = 4.33YY25 pKa = 11.28DD26 pKa = 4.37DD27 pKa = 4.9GATAFVQLPEE37 pKa = 4.38GGIDD41 pKa = 3.5ALLSGDD47 pKa = 4.46SPLAAPGHH55 pKa = 6.43GYY57 pKa = 10.04VPPQITATPPAHH69 pKa = 7.47DD70 pKa = 4.26GTDD73 pKa = 3.5PAAGAWGAPAGGTGWPDD90 pKa = 3.52PNAVPAQEE98 pKa = 4.6PGAGDD103 pKa = 3.56RR104 pKa = 11.84FTYY107 pKa = 10.44HH108 pKa = 7.21PGATGQWNFGDD119 pKa = 3.92ANGPEE124 pKa = 4.39PAPGPGHH131 pKa = 7.3DD132 pKa = 4.16VTGQWSIPVAGGDD145 pKa = 3.84LPDD148 pKa = 3.73EE149 pKa = 4.58SGEE152 pKa = 4.18YY153 pKa = 8.72STSALLEE160 pKa = 4.13QWGGTPPATLPGGAPAPWATGPGGAGGEE188 pKa = 4.27PWGQTAPSADD198 pKa = 4.65PYY200 pKa = 10.53AQPPGPYY207 pKa = 9.88DD208 pKa = 3.2SGAPGLPGGDD218 pKa = 3.0AHH220 pKa = 7.24GGPSPAAGAAEE231 pKa = 4.1AEE233 pKa = 4.67RR234 pKa = 11.84YY235 pKa = 9.79ASAADD240 pKa = 3.47AALDD244 pKa = 3.93EE245 pKa = 4.44AHH247 pKa = 6.34TAAEE251 pKa = 4.31ASSTAAVYY259 pKa = 7.47EE260 pKa = 4.41TFTGSPALPGATDD273 pKa = 3.16AHH275 pKa = 6.76AGHH278 pKa = 6.77GPGYY282 pKa = 10.35GRR284 pKa = 11.84QSEE287 pKa = 4.3QTFGQGVEE295 pKa = 4.51FTFGEE300 pKa = 4.53SGAEE304 pKa = 3.81QPPRR308 pKa = 11.84ASGAGTAYY316 pKa = 8.9DD317 pKa = 3.88TAPGSAYY324 pKa = 10.19DD325 pKa = 4.04SPSGLPEE332 pKa = 3.97EE333 pKa = 4.85ASSGFPDD340 pKa = 3.88APTDD344 pKa = 3.88GFPDD348 pKa = 4.05TPASSTEE355 pKa = 3.75QTEE358 pKa = 4.35AQQEE362 pKa = 4.54EE363 pKa = 5.19PGTPHH368 pKa = 6.85GSVSGVADD376 pKa = 4.02ATPSATDD383 pKa = 3.72GSTSGPEE390 pKa = 3.65GDD392 pKa = 4.81APPGFPGEE400 pKa = 4.31SALGAEE406 pKa = 5.06GPEE409 pKa = 4.03EE410 pKa = 4.24GAPSGAADD418 pKa = 4.32GSDD421 pKa = 3.28SGADD425 pKa = 3.22AAVTADD431 pKa = 4.13FPDD434 pKa = 3.67GRR436 pKa = 11.84APGDD440 pKa = 3.38GDD442 pKa = 3.77GTPEE446 pKa = 4.27GLPDD450 pKa = 4.11GAASDD455 pKa = 4.47ASPPLPPEE463 pKa = 4.2RR464 pKa = 11.84PAAAEE469 pKa = 3.67DD470 pKa = 3.47TGANEE475 pKa = 4.27EE476 pKa = 4.32SPEE479 pKa = 3.99YY480 pKa = 10.03PEE482 pKa = 4.86SRR484 pKa = 11.84EE485 pKa = 4.12SQASSASPDD494 pKa = 3.64GVPDD498 pKa = 3.45ATDD501 pKa = 3.22VVDD504 pKa = 5.28ASDD507 pKa = 4.28GADD510 pKa = 3.49PAPGDD515 pKa = 3.78GADD518 pKa = 4.56PSPEE522 pKa = 4.04PAPAAVPDD530 pKa = 3.74AATPHH535 pKa = 7.06DD536 pKa = 4.77APAPPQEE543 pKa = 4.05EE544 pKa = 4.67HH545 pKa = 6.88PLASYY550 pKa = 9.16VLRR553 pKa = 11.84VNGVDD558 pKa = 4.84RR559 pKa = 11.84PVTDD563 pKa = 2.96AWIGEE568 pKa = 4.15SLLYY572 pKa = 10.18VLRR575 pKa = 11.84EE576 pKa = 3.94RR577 pKa = 11.84LGLAGAKK584 pKa = 9.72DD585 pKa = 3.75GCSQGEE591 pKa = 4.12CGACNVQVDD600 pKa = 3.94GRR602 pKa = 11.84LVASCLVPSVTAAGSEE618 pKa = 4.0VRR620 pKa = 11.84TVEE623 pKa = 3.85GLAVDD628 pKa = 4.6GRR630 pKa = 11.84PSDD633 pKa = 3.66VQRR636 pKa = 11.84ALARR640 pKa = 11.84CGAVQCGFCVPGMAMTVHH658 pKa = 7.27DD659 pKa = 5.16LLEE662 pKa = 4.78GNPAPTEE669 pKa = 4.08LEE671 pKa = 4.08TRR673 pKa = 11.84QALCGNLCRR682 pKa = 11.84CSGYY686 pKa = 10.36RR687 pKa = 11.84GVLEE691 pKa = 3.9AVKK694 pKa = 10.18EE695 pKa = 4.37VVAEE699 pKa = 4.43RR700 pKa = 11.84EE701 pKa = 3.97AHH703 pKa = 5.13TAADD707 pKa = 4.16GEE709 pKa = 4.63TDD711 pKa = 3.16TGEE714 pKa = 4.3ARR716 pKa = 11.84IPHH719 pKa = 5.39QAGPGAGGVHH729 pKa = 6.81PSAFDD734 pKa = 3.46SPGFPSANGPSGPQGHH750 pKa = 6.63TDD752 pKa = 2.52SSGPHH757 pKa = 6.08DD758 pKa = 3.55QAYY761 pKa = 9.56GQDD764 pKa = 3.54GGQAA768 pKa = 3.33
Molecular weight: 75.66 kDa
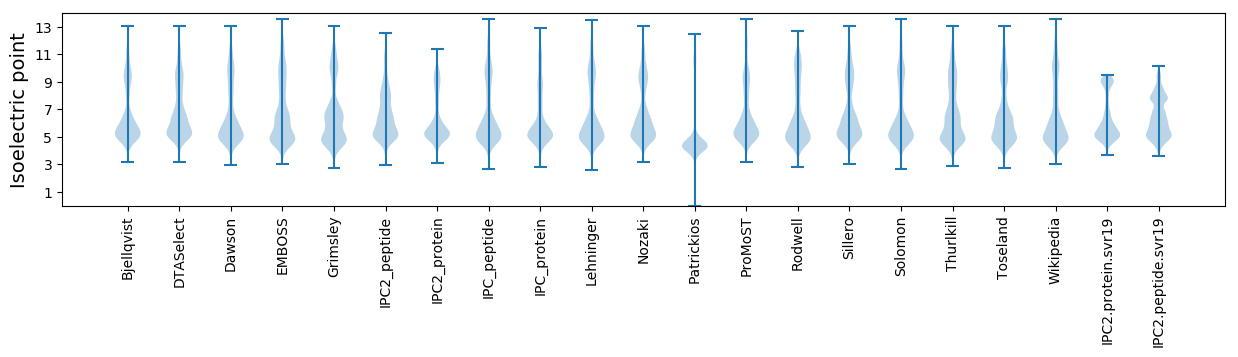
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R7HNJ8|A0A4R7HNJ8_9ACTN DNA-binding transcriptional LysR family regulator OS=Streptomyces sp. BK161 OX=887463 GN=EV573_11571 PE=3 SV=1
MM1 pKa = 7.55TGPPRR6 pKa = 11.84TSYY9 pKa = 10.4RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84NPGSPRR19 pKa = 11.84HH20 pKa = 5.98GPGPPPRR27 pKa = 11.84PVPRR31 pKa = 11.84RR32 pKa = 11.84APEE35 pKa = 4.2RR36 pKa = 11.84PHH38 pKa = 8.16ADD40 pKa = 3.23APPGNPRR47 pKa = 11.84PTHH50 pKa = 5.57PRR52 pKa = 11.84APRR55 pKa = 11.84ASHH58 pKa = 6.81PAPSPRR64 pKa = 11.84ASTSEE69 pKa = 3.91HH70 pKa = 5.66PTTRR74 pKa = 11.84LRR76 pKa = 11.84APPTASLRR84 pKa = 11.84APPTVPLRR92 pKa = 11.84ARR94 pKa = 11.84PATSPRR100 pKa = 11.84RR101 pKa = 11.84PPATSPRR108 pKa = 11.84PPPTAVRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84MGRR120 pKa = 11.84PRR122 pKa = 11.84SLGSVSRR129 pKa = 11.84AVVRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84MGRR138 pKa = 11.84PPASGPLSRR147 pKa = 11.84VVGRR151 pKa = 11.84RR152 pKa = 11.84RR153 pKa = 11.84MGRR156 pKa = 11.84PPASGLPSRR165 pKa = 11.84AVGRR169 pKa = 11.84RR170 pKa = 11.84PSRR173 pKa = 11.84HH174 pKa = 5.21SPSRR178 pKa = 11.84RR179 pKa = 11.84PRR181 pKa = 11.84SRR183 pKa = 11.84AVGRR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84MRR191 pKa = 11.84RR192 pKa = 11.84PPASGPRR199 pKa = 11.84SPAVGRR205 pKa = 11.84RR206 pKa = 11.84PTRR209 pKa = 11.84YY210 pKa = 8.74PPSPRR215 pKa = 11.84PPSRR219 pKa = 11.84VAVRR223 pKa = 11.84RR224 pKa = 11.84PMRR227 pKa = 11.84PQSPGPLSRR236 pKa = 11.84AVGRR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84MGRR245 pKa = 11.84PPAPGLLSRR254 pKa = 11.84AVGRR258 pKa = 11.84RR259 pKa = 11.84PSRR262 pKa = 11.84HH263 pKa = 5.97PPSPRR268 pKa = 11.84PPSRR272 pKa = 11.84AVGRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84MGRR281 pKa = 11.84PPASGLPSRR290 pKa = 11.84VAVRR294 pKa = 11.84RR295 pKa = 11.84PMRR298 pKa = 11.84PQSPGPLSRR307 pKa = 11.84VVGRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84MGRR316 pKa = 11.84PPAPGLTGRR325 pKa = 11.84AVGRR329 pKa = 11.84RR330 pKa = 11.84PMRR333 pKa = 11.84RR334 pKa = 11.84PPSPRR339 pKa = 11.84PPSRR343 pKa = 11.84AVVCRR348 pKa = 11.84PMRR351 pKa = 11.84RR352 pKa = 11.84PGPRR356 pKa = 11.84SIRR359 pKa = 11.84PTAARR364 pKa = 11.84RR365 pKa = 11.84TGVRR369 pKa = 11.84APQPRR374 pKa = 11.84VPPVIEE380 pKa = 4.65RR381 pKa = 11.84PWGHH385 pKa = 7.02PPQPLPSAPPTTPPP399 pKa = 3.56
MM1 pKa = 7.55TGPPRR6 pKa = 11.84TSYY9 pKa = 10.4RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84NPGSPRR19 pKa = 11.84HH20 pKa = 5.98GPGPPPRR27 pKa = 11.84PVPRR31 pKa = 11.84RR32 pKa = 11.84APEE35 pKa = 4.2RR36 pKa = 11.84PHH38 pKa = 8.16ADD40 pKa = 3.23APPGNPRR47 pKa = 11.84PTHH50 pKa = 5.57PRR52 pKa = 11.84APRR55 pKa = 11.84ASHH58 pKa = 6.81PAPSPRR64 pKa = 11.84ASTSEE69 pKa = 3.91HH70 pKa = 5.66PTTRR74 pKa = 11.84LRR76 pKa = 11.84APPTASLRR84 pKa = 11.84APPTVPLRR92 pKa = 11.84ARR94 pKa = 11.84PATSPRR100 pKa = 11.84RR101 pKa = 11.84PPATSPRR108 pKa = 11.84PPPTAVRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84MGRR120 pKa = 11.84PRR122 pKa = 11.84SLGSVSRR129 pKa = 11.84AVVRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84MGRR138 pKa = 11.84PPASGPLSRR147 pKa = 11.84VVGRR151 pKa = 11.84RR152 pKa = 11.84RR153 pKa = 11.84MGRR156 pKa = 11.84PPASGLPSRR165 pKa = 11.84AVGRR169 pKa = 11.84RR170 pKa = 11.84PSRR173 pKa = 11.84HH174 pKa = 5.21SPSRR178 pKa = 11.84RR179 pKa = 11.84PRR181 pKa = 11.84SRR183 pKa = 11.84AVGRR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84MRR191 pKa = 11.84RR192 pKa = 11.84PPASGPRR199 pKa = 11.84SPAVGRR205 pKa = 11.84RR206 pKa = 11.84PTRR209 pKa = 11.84YY210 pKa = 8.74PPSPRR215 pKa = 11.84PPSRR219 pKa = 11.84VAVRR223 pKa = 11.84RR224 pKa = 11.84PMRR227 pKa = 11.84PQSPGPLSRR236 pKa = 11.84AVGRR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84MGRR245 pKa = 11.84PPAPGLLSRR254 pKa = 11.84AVGRR258 pKa = 11.84RR259 pKa = 11.84PSRR262 pKa = 11.84HH263 pKa = 5.97PPSPRR268 pKa = 11.84PPSRR272 pKa = 11.84AVGRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84MGRR281 pKa = 11.84PPASGLPSRR290 pKa = 11.84VAVRR294 pKa = 11.84RR295 pKa = 11.84PMRR298 pKa = 11.84PQSPGPLSRR307 pKa = 11.84VVGRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84MGRR316 pKa = 11.84PPAPGLTGRR325 pKa = 11.84AVGRR329 pKa = 11.84RR330 pKa = 11.84PMRR333 pKa = 11.84RR334 pKa = 11.84PPSPRR339 pKa = 11.84PPSRR343 pKa = 11.84AVVCRR348 pKa = 11.84PMRR351 pKa = 11.84RR352 pKa = 11.84PGPRR356 pKa = 11.84SIRR359 pKa = 11.84PTAARR364 pKa = 11.84RR365 pKa = 11.84TGVRR369 pKa = 11.84APQPRR374 pKa = 11.84VPPVIEE380 pKa = 4.65RR381 pKa = 11.84PWGHH385 pKa = 7.02PPQPLPSAPPTTPPP399 pKa = 3.56
Molecular weight: 43.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3064903 |
25 |
8389 |
332.4 |
35.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.286 ± 0.035 | 0.802 ± 0.009 |
6.043 ± 0.022 | 5.647 ± 0.027 |
2.731 ± 0.015 | 9.349 ± 0.025 |
2.337 ± 0.013 | 3.095 ± 0.017 |
2.208 ± 0.024 | 10.277 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.691 ± 0.01 | 1.859 ± 0.016 |
6.107 ± 0.026 | 2.818 ± 0.015 |
7.98 ± 0.033 | 5.177 ± 0.019 |
6.344 ± 0.027 | 8.511 ± 0.023 |
1.581 ± 0.011 | 2.159 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
