
Lachnoclostridium sp. An169
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; unclassified Lachnoclostridium
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
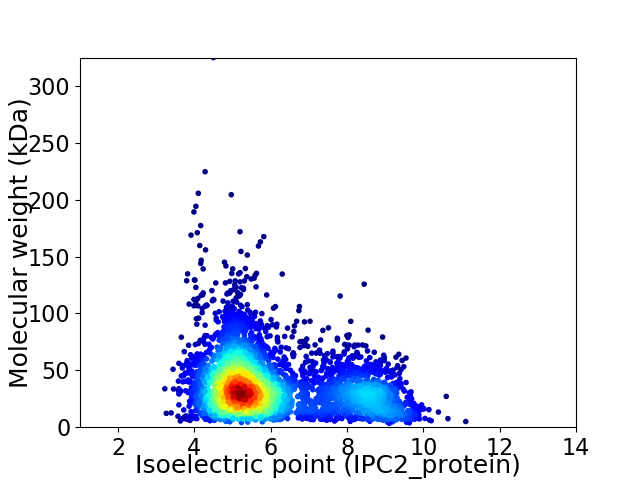
Virtual 2D-PAGE plot for 4280 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
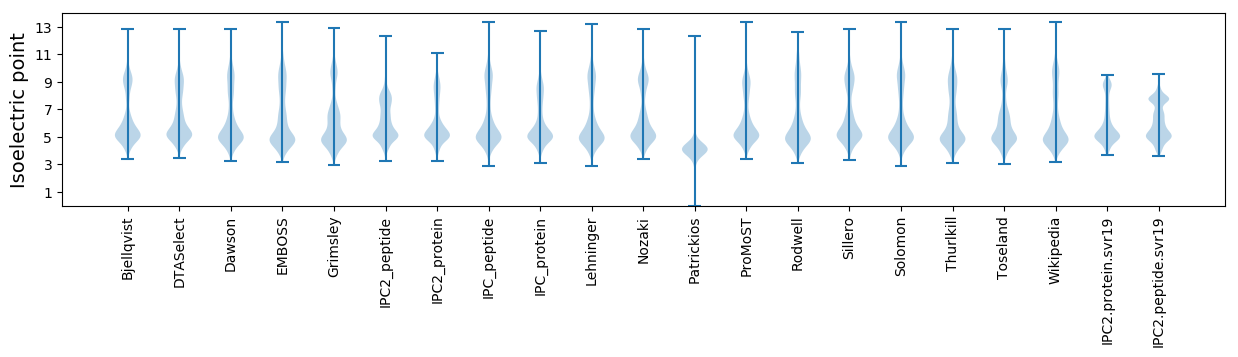
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4NJX2|A0A1Y4NJX2_9FIRM Cytidylate kinase OS=Lachnoclostridium sp. An169 OX=1965569 GN=B5F07_18555 PE=4 SV=1
MM1 pKa = 8.67KK2 pKa = 9.83MMKK5 pKa = 9.95KK6 pKa = 9.87AASVALASALVVSLAACGSNGGDD29 pKa = 3.48GNSNSGSTGSDD40 pKa = 2.28TFMIGGIGPTTGDD53 pKa = 3.02NAIYY57 pKa = 8.55GTAVKK62 pKa = 10.63NGIQLAVDD70 pKa = 4.95EE71 pKa = 4.7INEE74 pKa = 4.12AGGINGYY81 pKa = 9.16QISYY85 pKa = 10.25KK86 pKa = 10.59FEE88 pKa = 5.67DD89 pKa = 4.18DD90 pKa = 3.5QSDD93 pKa = 3.86SEE95 pKa = 4.59KK96 pKa = 11.03SVNAYY101 pKa = 7.69NTLKK105 pKa = 10.75DD106 pKa = 2.94WGMQMLVGTVTSTPCVAVVEE126 pKa = 4.56EE127 pKa = 4.2THH129 pKa = 6.17VDD131 pKa = 2.97NMFQFTPSATSVEE144 pKa = 4.32SVQYY148 pKa = 11.31DD149 pKa = 3.34NAFRR153 pKa = 11.84MCFSDD158 pKa = 4.25PSQGTVSADD167 pKa = 3.43YY168 pKa = 10.88IADD171 pKa = 3.56NGLATKK177 pKa = 10.3IGVIYY182 pKa = 10.15DD183 pKa = 3.45SSSTYY188 pKa = 9.52STGIYY193 pKa = 10.11QNFAAEE199 pKa = 4.23ADD201 pKa = 4.07VKK203 pKa = 11.06GLEE206 pKa = 4.22IVSAEE211 pKa = 4.04AFTSDD216 pKa = 3.78SNSDD220 pKa = 3.8FNVQLQKK227 pKa = 11.28AKK229 pKa = 10.65DD230 pKa = 3.4AGAEE234 pKa = 4.19LVFLPIYY241 pKa = 9.08YY242 pKa = 9.88QEE244 pKa = 4.49ASLILAQADD253 pKa = 3.63RR254 pKa = 11.84MGYY257 pKa = 8.06APKK260 pKa = 10.11WFGVDD265 pKa = 3.43GMDD268 pKa = 5.99GILNLDD274 pKa = 3.83GFDD277 pKa = 3.55ASLAEE282 pKa = 4.02GVMFLTPFTPTADD295 pKa = 4.21DD296 pKa = 3.84EE297 pKa = 4.45ATQTFVANYY306 pKa = 7.96EE307 pKa = 4.2AEE309 pKa = 4.4FGDD312 pKa = 4.31TPIQFAADD320 pKa = 4.19AYY322 pKa = 10.7DD323 pKa = 3.49CLYY326 pKa = 10.42VIKK329 pKa = 10.49AAAEE333 pKa = 4.13KK334 pKa = 10.86AGITPDD340 pKa = 3.31MSVSDD345 pKa = 3.74ICDD348 pKa = 3.25AMKK351 pKa = 9.59TAMTEE356 pKa = 3.69ITVDD360 pKa = 3.43GLTGKK365 pKa = 10.38QITWGEE371 pKa = 3.93DD372 pKa = 3.37GEE374 pKa = 4.37PSKK377 pKa = 11.25EE378 pKa = 3.88PTVVVIHH385 pKa = 7.0DD386 pKa = 3.74GAYY389 pKa = 9.18TVMM392 pKa = 4.95
MM1 pKa = 8.67KK2 pKa = 9.83MMKK5 pKa = 9.95KK6 pKa = 9.87AASVALASALVVSLAACGSNGGDD29 pKa = 3.48GNSNSGSTGSDD40 pKa = 2.28TFMIGGIGPTTGDD53 pKa = 3.02NAIYY57 pKa = 8.55GTAVKK62 pKa = 10.63NGIQLAVDD70 pKa = 4.95EE71 pKa = 4.7INEE74 pKa = 4.12AGGINGYY81 pKa = 9.16QISYY85 pKa = 10.25KK86 pKa = 10.59FEE88 pKa = 5.67DD89 pKa = 4.18DD90 pKa = 3.5QSDD93 pKa = 3.86SEE95 pKa = 4.59KK96 pKa = 11.03SVNAYY101 pKa = 7.69NTLKK105 pKa = 10.75DD106 pKa = 2.94WGMQMLVGTVTSTPCVAVVEE126 pKa = 4.56EE127 pKa = 4.2THH129 pKa = 6.17VDD131 pKa = 2.97NMFQFTPSATSVEE144 pKa = 4.32SVQYY148 pKa = 11.31DD149 pKa = 3.34NAFRR153 pKa = 11.84MCFSDD158 pKa = 4.25PSQGTVSADD167 pKa = 3.43YY168 pKa = 10.88IADD171 pKa = 3.56NGLATKK177 pKa = 10.3IGVIYY182 pKa = 10.15DD183 pKa = 3.45SSSTYY188 pKa = 9.52STGIYY193 pKa = 10.11QNFAAEE199 pKa = 4.23ADD201 pKa = 4.07VKK203 pKa = 11.06GLEE206 pKa = 4.22IVSAEE211 pKa = 4.04AFTSDD216 pKa = 3.78SNSDD220 pKa = 3.8FNVQLQKK227 pKa = 11.28AKK229 pKa = 10.65DD230 pKa = 3.4AGAEE234 pKa = 4.19LVFLPIYY241 pKa = 9.08YY242 pKa = 9.88QEE244 pKa = 4.49ASLILAQADD253 pKa = 3.63RR254 pKa = 11.84MGYY257 pKa = 8.06APKK260 pKa = 10.11WFGVDD265 pKa = 3.43GMDD268 pKa = 5.99GILNLDD274 pKa = 3.83GFDD277 pKa = 3.55ASLAEE282 pKa = 4.02GVMFLTPFTPTADD295 pKa = 4.21DD296 pKa = 3.84EE297 pKa = 4.45ATQTFVANYY306 pKa = 7.96EE307 pKa = 4.2AEE309 pKa = 4.4FGDD312 pKa = 4.31TPIQFAADD320 pKa = 4.19AYY322 pKa = 10.7DD323 pKa = 3.49CLYY326 pKa = 10.42VIKK329 pKa = 10.49AAAEE333 pKa = 4.13KK334 pKa = 10.86AGITPDD340 pKa = 3.31MSVSDD345 pKa = 3.74ICDD348 pKa = 3.25AMKK351 pKa = 9.59TAMTEE356 pKa = 3.69ITVDD360 pKa = 3.43GLTGKK365 pKa = 10.38QITWGEE371 pKa = 3.93DD372 pKa = 3.37GEE374 pKa = 4.37PSKK377 pKa = 11.25EE378 pKa = 3.88PTVVVIHH385 pKa = 7.0DD386 pKa = 3.74GAYY389 pKa = 9.18TVMM392 pKa = 4.95
Molecular weight: 41.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4NMH4|A0A1Y4NMH4_9FIRM Secretion protein F OS=Lachnoclostridium sp. An169 OX=1965569 GN=B5F07_15575 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1444677 |
27 |
2981 |
337.5 |
37.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.669 ± 0.037 | 1.542 ± 0.016 |
5.71 ± 0.033 | 8.02 ± 0.042 |
4.138 ± 0.029 | 7.456 ± 0.032 |
1.735 ± 0.014 | 6.727 ± 0.037 |
5.863 ± 0.033 | 8.902 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.065 ± 0.018 | 3.947 ± 0.026 |
3.555 ± 0.02 | 3.134 ± 0.022 |
5.332 ± 0.036 | 5.724 ± 0.03 |
5.369 ± 0.033 | 6.789 ± 0.031 |
1.07 ± 0.014 | 4.252 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
