
Mus musculus polyomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 5.57
Get precalculated fractions of proteins
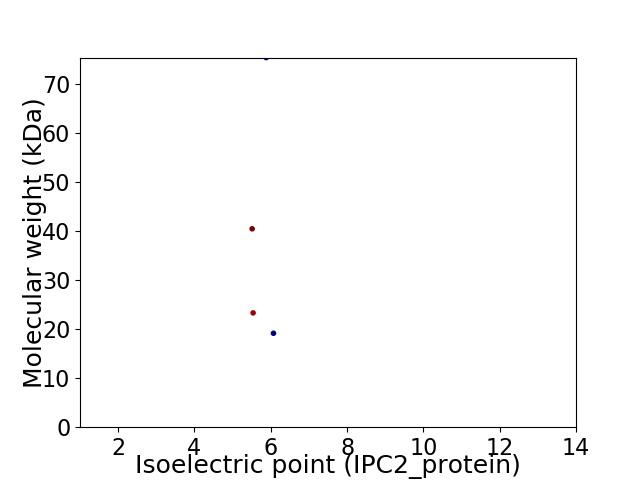
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A2IBB5|A2IBB5_9POLY Minor capsid protein OS=Mus musculus polyomavirus 2 OX=1891770 GN=VP3 PE=3 SV=1
MM1 pKa = 7.63APTVKK6 pKa = 9.86KK7 pKa = 10.62RR8 pKa = 11.84SVAKK12 pKa = 9.86PRR14 pKa = 11.84PKK16 pKa = 9.99PSEE19 pKa = 4.01VPKK22 pKa = 10.63LIVVGGIQVLDD33 pKa = 3.72VRR35 pKa = 11.84TGPDD39 pKa = 3.32SITQIEE45 pKa = 4.54AFLNPRR51 pKa = 11.84MGQPVDD57 pKa = 3.17SDD59 pKa = 4.17FYY61 pKa = 11.45GFSDD65 pKa = 4.82NITVSADD72 pKa = 3.22YY73 pKa = 8.99TQDD76 pKa = 2.91MPRR79 pKa = 11.84IKK81 pKa = 10.19EE82 pKa = 4.05LPCYY86 pKa = 10.78SMAKK90 pKa = 9.84ISLPMLNEE98 pKa = 4.64DD99 pKa = 3.95MTCDD103 pKa = 3.74TILMWEE109 pKa = 4.8AISCKK114 pKa = 9.66TEE116 pKa = 3.86VVGVSSLTNCHH127 pKa = 5.87SAVKK131 pKa = 10.12RR132 pKa = 11.84LYY134 pKa = 10.45DD135 pKa = 3.48NEE137 pKa = 4.36GAGFPVQGLNFHH149 pKa = 6.83FFSVGGEE156 pKa = 3.94ALDD159 pKa = 3.56LQMVVEE165 pKa = 5.05NYY167 pKa = 9.11RR168 pKa = 11.84CKK170 pKa = 10.91YY171 pKa = 7.66PAGVAALQAAPKK183 pKa = 9.3AAQVLDD189 pKa = 4.09PKK191 pKa = 11.02LKK193 pKa = 10.87AKK195 pKa = 9.1LTADD199 pKa = 3.18GKK201 pKa = 11.03FPIEE205 pKa = 4.08AWSPDD210 pKa = 3.33PAKK213 pKa = 10.98NEE215 pKa = 3.57NTRR218 pKa = 11.84YY219 pKa = 9.81FGTYY223 pKa = 8.95TGGLQTPPVLQITNTTTTILLNEE246 pKa = 4.48NGVGPLCKK254 pKa = 10.17GDD256 pKa = 3.96GLYY259 pKa = 10.57LASADD264 pKa = 2.93IVGFRR269 pKa = 11.84TQQNNKK275 pKa = 5.22MHH277 pKa = 6.34FRR279 pKa = 11.84GLPRR283 pKa = 11.84YY284 pKa = 9.57FSITLRR290 pKa = 11.84KK291 pKa = 9.53RR292 pKa = 11.84LVKK295 pKa = 10.54NPYY298 pKa = 9.07PVSSLLNTLFSEE310 pKa = 4.5MMPQIDD316 pKa = 4.07GQKK319 pKa = 9.32MEE321 pKa = 5.04GEE323 pKa = 4.23DD324 pKa = 3.56AQVEE328 pKa = 4.36EE329 pKa = 4.05VRR331 pKa = 11.84IYY333 pKa = 11.17DD334 pKa = 3.6GVEE337 pKa = 3.91RR338 pKa = 11.84LPGDD342 pKa = 3.64PDD344 pKa = 3.65MIRR347 pKa = 11.84YY348 pKa = 8.92RR349 pKa = 11.84DD350 pKa = 3.45KK351 pKa = 10.71FGQEE355 pKa = 3.53VTVPDD360 pKa = 3.82IPSNEE365 pKa = 3.83AA366 pKa = 2.98
MM1 pKa = 7.63APTVKK6 pKa = 9.86KK7 pKa = 10.62RR8 pKa = 11.84SVAKK12 pKa = 9.86PRR14 pKa = 11.84PKK16 pKa = 9.99PSEE19 pKa = 4.01VPKK22 pKa = 10.63LIVVGGIQVLDD33 pKa = 3.72VRR35 pKa = 11.84TGPDD39 pKa = 3.32SITQIEE45 pKa = 4.54AFLNPRR51 pKa = 11.84MGQPVDD57 pKa = 3.17SDD59 pKa = 4.17FYY61 pKa = 11.45GFSDD65 pKa = 4.82NITVSADD72 pKa = 3.22YY73 pKa = 8.99TQDD76 pKa = 2.91MPRR79 pKa = 11.84IKK81 pKa = 10.19EE82 pKa = 4.05LPCYY86 pKa = 10.78SMAKK90 pKa = 9.84ISLPMLNEE98 pKa = 4.64DD99 pKa = 3.95MTCDD103 pKa = 3.74TILMWEE109 pKa = 4.8AISCKK114 pKa = 9.66TEE116 pKa = 3.86VVGVSSLTNCHH127 pKa = 5.87SAVKK131 pKa = 10.12RR132 pKa = 11.84LYY134 pKa = 10.45DD135 pKa = 3.48NEE137 pKa = 4.36GAGFPVQGLNFHH149 pKa = 6.83FFSVGGEE156 pKa = 3.94ALDD159 pKa = 3.56LQMVVEE165 pKa = 5.05NYY167 pKa = 9.11RR168 pKa = 11.84CKK170 pKa = 10.91YY171 pKa = 7.66PAGVAALQAAPKK183 pKa = 9.3AAQVLDD189 pKa = 4.09PKK191 pKa = 11.02LKK193 pKa = 10.87AKK195 pKa = 9.1LTADD199 pKa = 3.18GKK201 pKa = 11.03FPIEE205 pKa = 4.08AWSPDD210 pKa = 3.33PAKK213 pKa = 10.98NEE215 pKa = 3.57NTRR218 pKa = 11.84YY219 pKa = 9.81FGTYY223 pKa = 8.95TGGLQTPPVLQITNTTTTILLNEE246 pKa = 4.48NGVGPLCKK254 pKa = 10.17GDD256 pKa = 3.96GLYY259 pKa = 10.57LASADD264 pKa = 2.93IVGFRR269 pKa = 11.84TQQNNKK275 pKa = 5.22MHH277 pKa = 6.34FRR279 pKa = 11.84GLPRR283 pKa = 11.84YY284 pKa = 9.57FSITLRR290 pKa = 11.84KK291 pKa = 9.53RR292 pKa = 11.84LVKK295 pKa = 10.54NPYY298 pKa = 9.07PVSSLLNTLFSEE310 pKa = 4.5MMPQIDD316 pKa = 4.07GQKK319 pKa = 9.32MEE321 pKa = 5.04GEE323 pKa = 4.23DD324 pKa = 3.56AQVEE328 pKa = 4.36EE329 pKa = 4.05VRR331 pKa = 11.84IYY333 pKa = 11.17DD334 pKa = 3.6GVEE337 pKa = 3.91RR338 pKa = 11.84LPGDD342 pKa = 3.64PDD344 pKa = 3.65MIRR347 pKa = 11.84YY348 pKa = 8.92RR349 pKa = 11.84DD350 pKa = 3.45KK351 pKa = 10.71FGQEE355 pKa = 3.53VTVPDD360 pKa = 3.82IPSNEE365 pKa = 3.83AA366 pKa = 2.98
Molecular weight: 40.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A2IBB2|A2IBB2_9POLY Small t antigen OS=Mus musculus polyomavirus 2 OX=1891770 GN=tAg PE=4 SV=1
MM1 pKa = 7.81DD2 pKa = 4.18HH3 pKa = 6.6QLTRR7 pKa = 11.84EE8 pKa = 3.73EE9 pKa = 4.49SQRR12 pKa = 11.84LMHH15 pKa = 6.93LLKK18 pKa = 10.89LPMEE22 pKa = 4.45QYY24 pKa = 11.84GNFPLMRR31 pKa = 11.84KK32 pKa = 9.54AFLRR36 pKa = 11.84ACKK39 pKa = 9.54IVHH42 pKa = 6.61PDD44 pKa = 3.02KK45 pKa = 11.35GGSDD49 pKa = 3.77EE50 pKa = 5.34LSQEE54 pKa = 4.36LISLYY59 pKa = 10.33RR60 pKa = 11.84RR61 pKa = 11.84LEE63 pKa = 3.8EE64 pKa = 4.71SLPCLSTQDD73 pKa = 4.68FIEE76 pKa = 4.14TDD78 pKa = 2.93ILQIPSYY85 pKa = 9.06GTPEE89 pKa = 3.44WDD91 pKa = 3.42EE92 pKa = 3.5WWKK95 pKa = 10.81EE96 pKa = 3.78FNKK99 pKa = 10.86DD100 pKa = 3.04FDD102 pKa = 4.59LFCNEE107 pKa = 4.07AFDD110 pKa = 5.51RR111 pKa = 11.84SDD113 pKa = 5.36DD114 pKa = 3.95EE115 pKa = 5.69QEE117 pKa = 4.13PQPDD121 pKa = 3.53DD122 pKa = 3.73SAPSSSRR129 pKa = 11.84PPTPPGSQATPPKK142 pKa = 9.87KK143 pKa = 9.42KK144 pKa = 10.92AKK146 pKa = 9.08MDD148 pKa = 3.87SPNDD152 pKa = 3.51MPADD156 pKa = 3.4LMEE159 pKa = 4.54YY160 pKa = 9.82LSCAILSNKK169 pKa = 9.33TITCFLIYY177 pKa = 7.83TTLEE181 pKa = 3.97KK182 pKa = 10.95SEE184 pKa = 4.3LLYY187 pKa = 11.35NKK189 pKa = 10.09LSEE192 pKa = 4.24KK193 pKa = 10.15YY194 pKa = 9.15KK195 pKa = 9.79PRR197 pKa = 11.84FISRR201 pKa = 11.84HH202 pKa = 4.98KK203 pKa = 11.14YY204 pKa = 9.16EE205 pKa = 4.44GNSLIFIITGTKK217 pKa = 9.62HH218 pKa = 5.52RR219 pKa = 11.84VSAVFNYY226 pKa = 9.48CATYY230 pKa = 10.33CSVSFIVVKK239 pKa = 10.3GVIKK243 pKa = 9.98EE244 pKa = 4.04YY245 pKa = 10.34PLYY248 pKa = 10.78CHH250 pKa = 7.33LCVEE254 pKa = 5.15PYY256 pKa = 10.58SVLQEE261 pKa = 4.37SIEE264 pKa = 4.28GGLNSEE270 pKa = 4.78FFDD273 pKa = 4.78APEE276 pKa = 4.24DD277 pKa = 3.49AAKK280 pKa = 10.32NVNWVAISEE289 pKa = 4.34YY290 pKa = 10.72ALKK293 pKa = 10.55INCDD297 pKa = 4.12DD298 pKa = 3.99IYY300 pKa = 11.58LLMGLYY306 pKa = 10.09KK307 pKa = 10.25EE308 pKa = 4.59FQSPVPNCSKK318 pKa = 10.8CEE320 pKa = 3.79NRR322 pKa = 11.84MLTNHH327 pKa = 6.16FKK329 pKa = 10.46FHH331 pKa = 7.06KK332 pKa = 9.05EE333 pKa = 3.5HH334 pKa = 7.05HH335 pKa = 6.35EE336 pKa = 4.01NALLFAACRR345 pKa = 11.84NQKK348 pKa = 8.25TICQQAVDD356 pKa = 3.95GVIAQRR362 pKa = 11.84RR363 pKa = 11.84VDD365 pKa = 3.75MAQMTRR371 pKa = 11.84NEE373 pKa = 3.73QLAARR378 pKa = 11.84FDD380 pKa = 3.78KK381 pKa = 10.93LFEE384 pKa = 4.2RR385 pKa = 11.84LEE387 pKa = 4.62VILSAQSSYY396 pKa = 9.85TISMFMAGIVWFEE409 pKa = 3.82NLFPGQSFKK418 pKa = 11.48DD419 pKa = 3.53LLLEE423 pKa = 4.22LLEE426 pKa = 4.74CMVSNIPKK434 pKa = 9.07RR435 pKa = 11.84RR436 pKa = 11.84YY437 pKa = 8.45WLFTGPVNTGKK448 pKa = 7.5TTLAAAVLDD457 pKa = 4.08LCGGKK462 pKa = 9.79ALNINMPFDD471 pKa = 3.7KK472 pKa = 11.05LNFEE476 pKa = 4.93LGVAIDD482 pKa = 3.29QFMVVFEE489 pKa = 4.43DD490 pKa = 3.6VKK492 pKa = 11.02GQKK495 pKa = 9.5SEE497 pKa = 4.59NKK499 pKa = 9.81DD500 pKa = 3.53LPPGQGINNLDD511 pKa = 3.91NLRR514 pKa = 11.84DD515 pKa = 3.9HH516 pKa = 7.37LDD518 pKa = 3.48GAVKK522 pKa = 10.63VNLEE526 pKa = 4.2KK527 pKa = 10.85KK528 pKa = 9.73HH529 pKa = 6.21LNKK532 pKa = 9.17KK533 pKa = 6.38TQIFPPGIVTCNEE546 pKa = 3.51YY547 pKa = 10.7FIPFTLRR554 pKa = 11.84VRR556 pKa = 11.84FCKK559 pKa = 10.5KK560 pKa = 10.3LVFKK564 pKa = 10.48FSKK567 pKa = 10.14YY568 pKa = 9.87QYY570 pKa = 10.83LSLKK574 pKa = 9.16KK575 pKa = 8.41TEE577 pKa = 3.95CLGRR581 pKa = 11.84YY582 pKa = 9.44RR583 pKa = 11.84ILQNGCTLLLLLIYY597 pKa = 10.57HH598 pKa = 7.26CDD600 pKa = 3.48LDD602 pKa = 5.24DD603 pKa = 4.13FAEE606 pKa = 4.59SIQGKK611 pKa = 8.22VRR613 pKa = 11.84AWKK616 pKa = 10.01EE617 pKa = 3.82RR618 pKa = 11.84VNSEE622 pKa = 3.92ISVSTYY628 pKa = 11.01LEE630 pKa = 3.87MRR632 pKa = 11.84QCCLEE637 pKa = 4.11GKK639 pKa = 10.19DD640 pKa = 3.43ILSARR645 pKa = 11.84NTQMPTQHH653 pKa = 6.52NEE655 pKa = 3.35
MM1 pKa = 7.81DD2 pKa = 4.18HH3 pKa = 6.6QLTRR7 pKa = 11.84EE8 pKa = 3.73EE9 pKa = 4.49SQRR12 pKa = 11.84LMHH15 pKa = 6.93LLKK18 pKa = 10.89LPMEE22 pKa = 4.45QYY24 pKa = 11.84GNFPLMRR31 pKa = 11.84KK32 pKa = 9.54AFLRR36 pKa = 11.84ACKK39 pKa = 9.54IVHH42 pKa = 6.61PDD44 pKa = 3.02KK45 pKa = 11.35GGSDD49 pKa = 3.77EE50 pKa = 5.34LSQEE54 pKa = 4.36LISLYY59 pKa = 10.33RR60 pKa = 11.84RR61 pKa = 11.84LEE63 pKa = 3.8EE64 pKa = 4.71SLPCLSTQDD73 pKa = 4.68FIEE76 pKa = 4.14TDD78 pKa = 2.93ILQIPSYY85 pKa = 9.06GTPEE89 pKa = 3.44WDD91 pKa = 3.42EE92 pKa = 3.5WWKK95 pKa = 10.81EE96 pKa = 3.78FNKK99 pKa = 10.86DD100 pKa = 3.04FDD102 pKa = 4.59LFCNEE107 pKa = 4.07AFDD110 pKa = 5.51RR111 pKa = 11.84SDD113 pKa = 5.36DD114 pKa = 3.95EE115 pKa = 5.69QEE117 pKa = 4.13PQPDD121 pKa = 3.53DD122 pKa = 3.73SAPSSSRR129 pKa = 11.84PPTPPGSQATPPKK142 pKa = 9.87KK143 pKa = 9.42KK144 pKa = 10.92AKK146 pKa = 9.08MDD148 pKa = 3.87SPNDD152 pKa = 3.51MPADD156 pKa = 3.4LMEE159 pKa = 4.54YY160 pKa = 9.82LSCAILSNKK169 pKa = 9.33TITCFLIYY177 pKa = 7.83TTLEE181 pKa = 3.97KK182 pKa = 10.95SEE184 pKa = 4.3LLYY187 pKa = 11.35NKK189 pKa = 10.09LSEE192 pKa = 4.24KK193 pKa = 10.15YY194 pKa = 9.15KK195 pKa = 9.79PRR197 pKa = 11.84FISRR201 pKa = 11.84HH202 pKa = 4.98KK203 pKa = 11.14YY204 pKa = 9.16EE205 pKa = 4.44GNSLIFIITGTKK217 pKa = 9.62HH218 pKa = 5.52RR219 pKa = 11.84VSAVFNYY226 pKa = 9.48CATYY230 pKa = 10.33CSVSFIVVKK239 pKa = 10.3GVIKK243 pKa = 9.98EE244 pKa = 4.04YY245 pKa = 10.34PLYY248 pKa = 10.78CHH250 pKa = 7.33LCVEE254 pKa = 5.15PYY256 pKa = 10.58SVLQEE261 pKa = 4.37SIEE264 pKa = 4.28GGLNSEE270 pKa = 4.78FFDD273 pKa = 4.78APEE276 pKa = 4.24DD277 pKa = 3.49AAKK280 pKa = 10.32NVNWVAISEE289 pKa = 4.34YY290 pKa = 10.72ALKK293 pKa = 10.55INCDD297 pKa = 4.12DD298 pKa = 3.99IYY300 pKa = 11.58LLMGLYY306 pKa = 10.09KK307 pKa = 10.25EE308 pKa = 4.59FQSPVPNCSKK318 pKa = 10.8CEE320 pKa = 3.79NRR322 pKa = 11.84MLTNHH327 pKa = 6.16FKK329 pKa = 10.46FHH331 pKa = 7.06KK332 pKa = 9.05EE333 pKa = 3.5HH334 pKa = 7.05HH335 pKa = 6.35EE336 pKa = 4.01NALLFAACRR345 pKa = 11.84NQKK348 pKa = 8.25TICQQAVDD356 pKa = 3.95GVIAQRR362 pKa = 11.84RR363 pKa = 11.84VDD365 pKa = 3.75MAQMTRR371 pKa = 11.84NEE373 pKa = 3.73QLAARR378 pKa = 11.84FDD380 pKa = 3.78KK381 pKa = 10.93LFEE384 pKa = 4.2RR385 pKa = 11.84LEE387 pKa = 4.62VILSAQSSYY396 pKa = 9.85TISMFMAGIVWFEE409 pKa = 3.82NLFPGQSFKK418 pKa = 11.48DD419 pKa = 3.53LLLEE423 pKa = 4.22LLEE426 pKa = 4.74CMVSNIPKK434 pKa = 9.07RR435 pKa = 11.84RR436 pKa = 11.84YY437 pKa = 8.45WLFTGPVNTGKK448 pKa = 7.5TTLAAAVLDD457 pKa = 4.08LCGGKK462 pKa = 9.79ALNINMPFDD471 pKa = 3.7KK472 pKa = 11.05LNFEE476 pKa = 4.93LGVAIDD482 pKa = 3.29QFMVVFEE489 pKa = 4.43DD490 pKa = 3.6VKK492 pKa = 11.02GQKK495 pKa = 9.5SEE497 pKa = 4.59NKK499 pKa = 9.81DD500 pKa = 3.53LPPGQGINNLDD511 pKa = 3.91NLRR514 pKa = 11.84DD515 pKa = 3.9HH516 pKa = 7.37LDD518 pKa = 3.48GAVKK522 pKa = 10.63VNLEE526 pKa = 4.2KK527 pKa = 10.85KK528 pKa = 9.73HH529 pKa = 6.21LNKK532 pKa = 9.17KK533 pKa = 6.38TQIFPPGIVTCNEE546 pKa = 3.51YY547 pKa = 10.7FIPFTLRR554 pKa = 11.84VRR556 pKa = 11.84FCKK559 pKa = 10.5KK560 pKa = 10.3LVFKK564 pKa = 10.48FSKK567 pKa = 10.14YY568 pKa = 9.87QYY570 pKa = 10.83LSLKK574 pKa = 9.16KK575 pKa = 8.41TEE577 pKa = 3.95CLGRR581 pKa = 11.84YY582 pKa = 9.44RR583 pKa = 11.84ILQNGCTLLLLLIYY597 pKa = 10.57HH598 pKa = 7.26CDD600 pKa = 3.48LDD602 pKa = 5.24DD603 pKa = 4.13FAEE606 pKa = 4.59SIQGKK611 pKa = 8.22VRR613 pKa = 11.84AWKK616 pKa = 10.01EE617 pKa = 3.82RR618 pKa = 11.84VNSEE622 pKa = 3.92ISVSTYY628 pKa = 11.01LEE630 pKa = 3.87MRR632 pKa = 11.84QCCLEE637 pKa = 4.11GKK639 pKa = 10.19DD640 pKa = 3.43ILSARR645 pKa = 11.84NTQMPTQHH653 pKa = 6.52NEE655 pKa = 3.35
Molecular weight: 75.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1386 |
160 |
655 |
346.5 |
39.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.916 ± 1.035 | 2.958 ± 0.977 |
5.628 ± 0.24 | 6.566 ± 0.502 |
4.69 ± 0.614 | 5.339 ± 0.893 |
1.804 ± 0.384 | 5.051 ± 0.547 |
6.349 ± 0.877 | 10.75 ± 0.959 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.886 ± 0.306 | 4.762 ± 0.513 |
5.916 ± 0.759 | 4.762 ± 0.533 |
4.834 ± 0.417 | 6.205 ± 0.387 |
4.762 ± 0.554 | 5.267 ± 0.998 |
1.443 ± 0.56 | 4.113 ± 0.611 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
