
Winogradskyella sediminis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
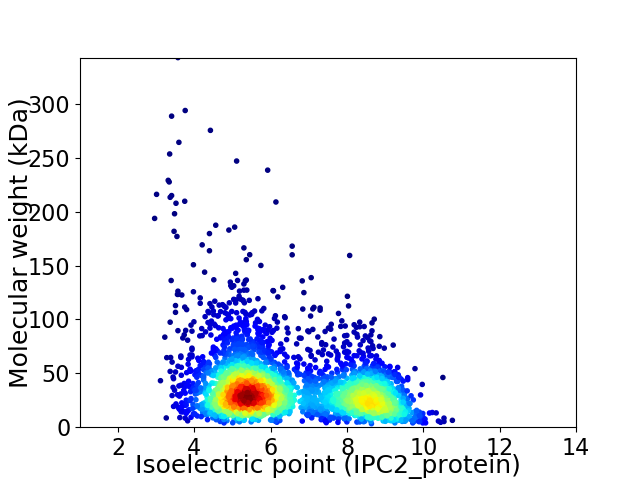
Virtual 2D-PAGE plot for 3162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1H1X232|A0A1H1X232_9FLAO Helix-turn-helix Psq domain OS=Winogradskyella sediminis OX=1382466 GN=SAMN04489797_3073 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.37RR3 pKa = 11.84LLSFSLCFGILLFWSSCRR21 pKa = 11.84NDD23 pKa = 3.92FEE25 pKa = 4.5FTPSKK30 pKa = 11.18GNLEE34 pKa = 4.2FAQDD38 pKa = 4.11TIYY41 pKa = 11.18LDD43 pKa = 3.74TVFTNIGSSTYY54 pKa = 9.91NLKK57 pKa = 10.64VYY59 pKa = 10.31NRR61 pKa = 11.84SDD63 pKa = 3.82DD64 pKa = 5.08DD65 pKa = 3.74IVIPTIQFEE74 pKa = 4.32NGVNSLYY81 pKa = 10.96RR82 pKa = 11.84MNIDD86 pKa = 3.31GNTGLEE92 pKa = 4.1DD93 pKa = 3.57AQEE96 pKa = 4.27GKK98 pKa = 10.61FFEE101 pKa = 5.1DD102 pKa = 4.23VEE104 pKa = 4.47LLANDD109 pKa = 3.69SLFIFIEE116 pKa = 4.18TTIDD120 pKa = 3.13ISTLGALEE128 pKa = 4.04NQFLYY133 pKa = 10.04TDD135 pKa = 4.99KK136 pKa = 11.12ILFDD140 pKa = 4.29SGNNQQDD147 pKa = 3.12IDD149 pKa = 4.18VVTLVKK155 pKa = 10.54DD156 pKa = 3.42AVFIYY161 pKa = 9.81PSKK164 pKa = 11.13DD165 pKa = 3.06DD166 pKa = 3.29TTGIVEE172 pKa = 4.3TLSFDD177 pKa = 3.28VDD179 pKa = 3.57GDD181 pKa = 3.93GTPDD185 pKa = 3.16EE186 pKa = 4.53TTLEE190 pKa = 4.27GRR192 pKa = 11.84FLTDD196 pKa = 4.93DD197 pKa = 3.62EE198 pKa = 4.75LTFTNEE204 pKa = 3.56KK205 pKa = 10.09PYY207 pKa = 10.86VIYY210 pKa = 10.07GYY212 pKa = 11.03AGVGSGKK219 pKa = 8.08TLTMEE224 pKa = 4.56AGARR228 pKa = 11.84VHH230 pKa = 6.27FHH232 pKa = 7.05ADD234 pKa = 2.64SGIIVTDD241 pKa = 3.63NASLQINGALSTDD254 pKa = 3.38QEE256 pKa = 4.41TLEE259 pKa = 4.44NEE261 pKa = 4.52VILEE265 pKa = 3.94GDD267 pKa = 3.35RR268 pKa = 11.84LEE270 pKa = 4.65PLYY273 pKa = 10.94EE274 pKa = 5.22DD275 pKa = 5.6IPGQWGTIWLYY286 pKa = 11.12NGSEE290 pKa = 4.23NNTINYY296 pKa = 7.49ATIKK300 pKa = 10.29NATIGVLSEE309 pKa = 4.38GNQNAPTDD317 pKa = 3.74KK318 pKa = 10.54LTITNTQIYY327 pKa = 8.86NVSSFGILGRR337 pKa = 11.84ATSIRR342 pKa = 11.84AEE344 pKa = 3.63NLVINNAGQSSFAGTYY360 pKa = 8.12GGKK363 pKa = 10.68YY364 pKa = 9.81NVTHH368 pKa = 5.44STIANYY374 pKa = 9.01WNSSFRR380 pKa = 11.84EE381 pKa = 4.1YY382 pKa = 10.3PALLINDD389 pKa = 4.62YY390 pKa = 10.99ILDD393 pKa = 3.91EE394 pKa = 4.67NNTIFTNDD402 pKa = 2.86LTEE405 pKa = 5.08ANFNNCIIYY414 pKa = 10.83GNDD417 pKa = 3.09NPEE420 pKa = 4.42FILEE424 pKa = 4.15NEE426 pKa = 4.02GDD428 pKa = 3.75VFNYY432 pKa = 10.37KK433 pKa = 8.62FTNCLLRR440 pKa = 11.84FNDD443 pKa = 4.72NNLQGNGNYY452 pKa = 10.27NFDD455 pKa = 3.6DD456 pKa = 3.57TAFYY460 pKa = 10.24IDD462 pKa = 4.1NVFNEE467 pKa = 4.38NPDD470 pKa = 3.45FEE472 pKa = 5.82DD473 pKa = 4.68PYY475 pKa = 10.87EE476 pKa = 4.53SLMKK480 pKa = 10.23IGEE483 pKa = 4.32DD484 pKa = 3.54SSAKK488 pKa = 10.43GIGSATYY495 pKa = 11.06SNLVLNDD502 pKa = 3.17ILGASRR508 pKa = 11.84IDD510 pKa = 3.47SGSFDD515 pKa = 3.07AGAYY519 pKa = 9.5NFSTFEE525 pKa = 4.1DD526 pKa = 3.59
MM1 pKa = 7.37KK2 pKa = 10.37RR3 pKa = 11.84LLSFSLCFGILLFWSSCRR21 pKa = 11.84NDD23 pKa = 3.92FEE25 pKa = 4.5FTPSKK30 pKa = 11.18GNLEE34 pKa = 4.2FAQDD38 pKa = 4.11TIYY41 pKa = 11.18LDD43 pKa = 3.74TVFTNIGSSTYY54 pKa = 9.91NLKK57 pKa = 10.64VYY59 pKa = 10.31NRR61 pKa = 11.84SDD63 pKa = 3.82DD64 pKa = 5.08DD65 pKa = 3.74IVIPTIQFEE74 pKa = 4.32NGVNSLYY81 pKa = 10.96RR82 pKa = 11.84MNIDD86 pKa = 3.31GNTGLEE92 pKa = 4.1DD93 pKa = 3.57AQEE96 pKa = 4.27GKK98 pKa = 10.61FFEE101 pKa = 5.1DD102 pKa = 4.23VEE104 pKa = 4.47LLANDD109 pKa = 3.69SLFIFIEE116 pKa = 4.18TTIDD120 pKa = 3.13ISTLGALEE128 pKa = 4.04NQFLYY133 pKa = 10.04TDD135 pKa = 4.99KK136 pKa = 11.12ILFDD140 pKa = 4.29SGNNQQDD147 pKa = 3.12IDD149 pKa = 4.18VVTLVKK155 pKa = 10.54DD156 pKa = 3.42AVFIYY161 pKa = 9.81PSKK164 pKa = 11.13DD165 pKa = 3.06DD166 pKa = 3.29TTGIVEE172 pKa = 4.3TLSFDD177 pKa = 3.28VDD179 pKa = 3.57GDD181 pKa = 3.93GTPDD185 pKa = 3.16EE186 pKa = 4.53TTLEE190 pKa = 4.27GRR192 pKa = 11.84FLTDD196 pKa = 4.93DD197 pKa = 3.62EE198 pKa = 4.75LTFTNEE204 pKa = 3.56KK205 pKa = 10.09PYY207 pKa = 10.86VIYY210 pKa = 10.07GYY212 pKa = 11.03AGVGSGKK219 pKa = 8.08TLTMEE224 pKa = 4.56AGARR228 pKa = 11.84VHH230 pKa = 6.27FHH232 pKa = 7.05ADD234 pKa = 2.64SGIIVTDD241 pKa = 3.63NASLQINGALSTDD254 pKa = 3.38QEE256 pKa = 4.41TLEE259 pKa = 4.44NEE261 pKa = 4.52VILEE265 pKa = 3.94GDD267 pKa = 3.35RR268 pKa = 11.84LEE270 pKa = 4.65PLYY273 pKa = 10.94EE274 pKa = 5.22DD275 pKa = 5.6IPGQWGTIWLYY286 pKa = 11.12NGSEE290 pKa = 4.23NNTINYY296 pKa = 7.49ATIKK300 pKa = 10.29NATIGVLSEE309 pKa = 4.38GNQNAPTDD317 pKa = 3.74KK318 pKa = 10.54LTITNTQIYY327 pKa = 8.86NVSSFGILGRR337 pKa = 11.84ATSIRR342 pKa = 11.84AEE344 pKa = 3.63NLVINNAGQSSFAGTYY360 pKa = 8.12GGKK363 pKa = 10.68YY364 pKa = 9.81NVTHH368 pKa = 5.44STIANYY374 pKa = 9.01WNSSFRR380 pKa = 11.84EE381 pKa = 4.1YY382 pKa = 10.3PALLINDD389 pKa = 4.62YY390 pKa = 10.99ILDD393 pKa = 3.91EE394 pKa = 4.67NNTIFTNDD402 pKa = 2.86LTEE405 pKa = 5.08ANFNNCIIYY414 pKa = 10.83GNDD417 pKa = 3.09NPEE420 pKa = 4.42FILEE424 pKa = 4.15NEE426 pKa = 4.02GDD428 pKa = 3.75VFNYY432 pKa = 10.37KK433 pKa = 8.62FTNCLLRR440 pKa = 11.84FNDD443 pKa = 4.72NNLQGNGNYY452 pKa = 10.27NFDD455 pKa = 3.6DD456 pKa = 3.57TAFYY460 pKa = 10.24IDD462 pKa = 4.1NVFNEE467 pKa = 4.38NPDD470 pKa = 3.45FEE472 pKa = 5.82DD473 pKa = 4.68PYY475 pKa = 10.87EE476 pKa = 4.53SLMKK480 pKa = 10.23IGEE483 pKa = 4.32DD484 pKa = 3.54SSAKK488 pKa = 10.43GIGSATYY495 pKa = 11.06SNLVLNDD502 pKa = 3.17ILGASRR508 pKa = 11.84IDD510 pKa = 3.47SGSFDD515 pKa = 3.07AGAYY519 pKa = 9.5NFSTFEE525 pKa = 4.1DD526 pKa = 3.59
Molecular weight: 58.5 kDa
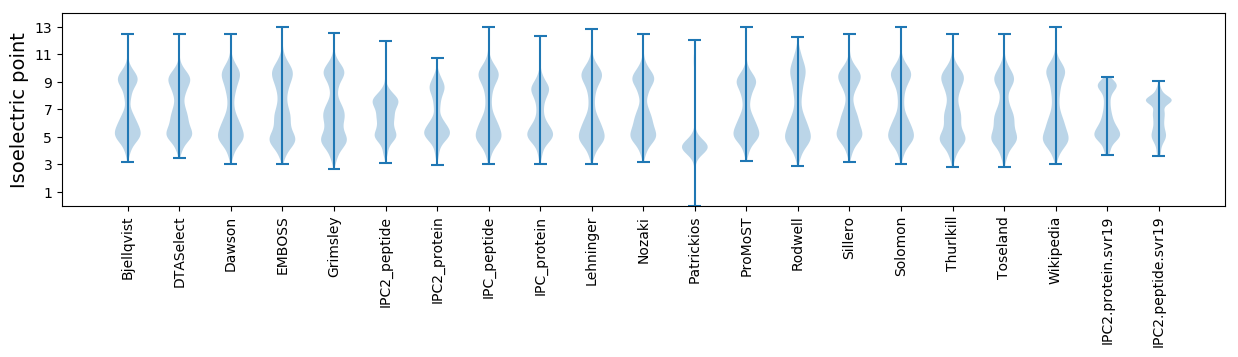
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1N9V9|A0A1H1N9V9_9FLAO Phosphoribosylformylglycinamidine synthase OS=Winogradskyella sediminis OX=1382466 GN=purL PE=3 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
Molecular weight: 6.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1098209 |
29 |
3263 |
347.3 |
39.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.44 ± 0.037 | 0.754 ± 0.016 |
5.875 ± 0.039 | 6.472 ± 0.042 |
5.14 ± 0.037 | 6.158 ± 0.041 |
1.835 ± 0.021 | 8.061 ± 0.041 |
7.417 ± 0.068 | 9.253 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.117 ± 0.021 | 6.338 ± 0.052 |
3.279 ± 0.028 | 3.333 ± 0.022 |
3.219 ± 0.031 | 6.667 ± 0.039 |
6.25 ± 0.061 | 6.243 ± 0.032 |
0.985 ± 0.015 | 4.166 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
