
Polyporus brumalis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporaceae; Polyporus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
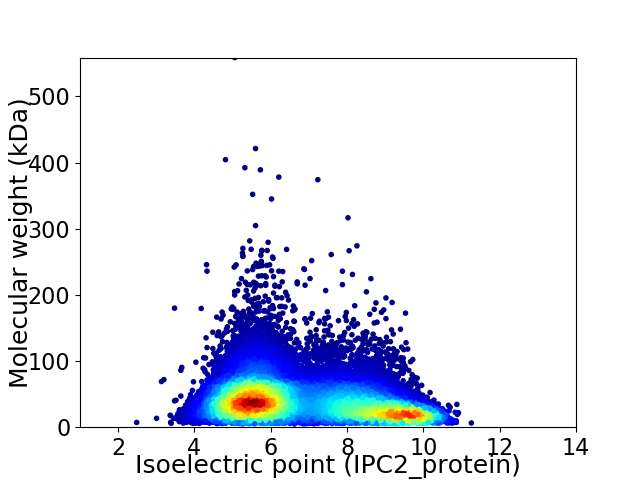
Virtual 2D-PAGE plot for 18174 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A371CGF6|A0A371CGF6_9APHY Uncharacterized protein (Fragment) OS=Polyporus brumalis OX=139420 GN=OH76DRAFT_1491264 PE=4 SV=1
MM1 pKa = 7.32EE2 pKa = 5.16AADD5 pKa = 3.76SGFVPMYY12 pKa = 10.38LDD14 pKa = 4.91SSPLDD19 pKa = 3.88DD20 pKa = 4.64AQGSGYY26 pKa = 10.56AEE28 pKa = 4.52CWDD31 pKa = 3.94SPRR34 pKa = 11.84ARR36 pKa = 11.84HH37 pKa = 6.1SVLANPYY44 pKa = 8.58KK45 pKa = 9.78PARR48 pKa = 11.84ISFYY52 pKa = 10.37PLSDD56 pKa = 3.34NMGSSLPPRR65 pKa = 11.84FASPGMPPLPSSHH78 pKa = 6.59PQLYY82 pKa = 9.05QQVDD86 pKa = 3.68TTFFCCSPEE95 pKa = 3.93PMSPQSQVAASFQLSTPSYY114 pKa = 10.97LEE116 pKa = 4.36LPPPLAGAEE125 pKa = 4.43FISDD129 pKa = 4.46LPTAQPALEE138 pKa = 4.36SMEE141 pKa = 4.85SPLPPSDD148 pKa = 3.79LSEE151 pKa = 4.5FSWHH155 pKa = 5.4TSSRR159 pKa = 11.84LLEE162 pKa = 4.52SDD164 pKa = 4.95DD165 pKa = 3.8VPQASFTMSPGEE177 pKa = 4.02SWIEE181 pKa = 3.63EE182 pKa = 4.24QPFGEE187 pKa = 4.91HH188 pKa = 7.81DD189 pKa = 4.83DD190 pKa = 3.98DD191 pKa = 4.39TSDD194 pKa = 3.15GSAYY198 pKa = 10.38SLPGGRR204 pKa = 11.84THH206 pKa = 8.19PILFDD211 pKa = 3.33EE212 pKa = 5.09AEE214 pKa = 4.33WNGATWDD221 pKa = 4.08AVCLADD227 pKa = 4.59FLGGLFEE234 pKa = 6.02KK235 pKa = 10.63ADD237 pKa = 3.64PWSTLDD243 pKa = 4.98DD244 pKa = 4.37FLDD247 pKa = 4.23VPSLLPDD254 pKa = 4.31LPPTSTSLFLLASDD268 pKa = 4.18RR269 pKa = 11.84GGVGYY274 pKa = 10.52VATPRR279 pKa = 11.84SPDD282 pKa = 3.18WQDD285 pKa = 3.76LSSQTLAEE293 pKa = 4.3EE294 pKa = 4.43TRR296 pKa = 11.84ACTPLFSVSCGTEE309 pKa = 3.58ALEE312 pKa = 5.12DD313 pKa = 3.93VPAVDD318 pKa = 4.96SDD320 pKa = 4.21PPSSGDD326 pKa = 3.14TAYY329 pKa = 10.99DD330 pKa = 3.56DD331 pKa = 4.2TPEE334 pKa = 3.97EE335 pKa = 4.05AQAYY339 pKa = 8.93LPRR342 pKa = 11.84AEE344 pKa = 4.62YY345 pKa = 10.59EE346 pKa = 4.19PLSAPAMPSPSPVEE360 pKa = 4.03MLVKK364 pKa = 10.37PPGVTVRR371 pKa = 11.84GPCASIPAAEE381 pKa = 4.52EE382 pKa = 4.17DD383 pKa = 3.83SDD385 pKa = 4.67VLEE388 pKa = 4.81EE389 pKa = 4.32KK390 pKa = 10.4CGHH393 pKa = 6.59DD394 pKa = 4.32ASLGTDD400 pKa = 3.42GADD403 pKa = 2.92AAVEE407 pKa = 4.07RR408 pKa = 11.84GIEE411 pKa = 4.06TGVPEE416 pKa = 4.47SFDD419 pKa = 3.7RR420 pKa = 11.84DD421 pKa = 3.65VSVDD425 pKa = 3.69GPSLFADD432 pKa = 4.41DD433 pKa = 6.37DD434 pKa = 4.27VDD436 pKa = 6.16LDD438 pKa = 4.75DD439 pKa = 4.61EE440 pKa = 4.53
MM1 pKa = 7.32EE2 pKa = 5.16AADD5 pKa = 3.76SGFVPMYY12 pKa = 10.38LDD14 pKa = 4.91SSPLDD19 pKa = 3.88DD20 pKa = 4.64AQGSGYY26 pKa = 10.56AEE28 pKa = 4.52CWDD31 pKa = 3.94SPRR34 pKa = 11.84ARR36 pKa = 11.84HH37 pKa = 6.1SVLANPYY44 pKa = 8.58KK45 pKa = 9.78PARR48 pKa = 11.84ISFYY52 pKa = 10.37PLSDD56 pKa = 3.34NMGSSLPPRR65 pKa = 11.84FASPGMPPLPSSHH78 pKa = 6.59PQLYY82 pKa = 9.05QQVDD86 pKa = 3.68TTFFCCSPEE95 pKa = 3.93PMSPQSQVAASFQLSTPSYY114 pKa = 10.97LEE116 pKa = 4.36LPPPLAGAEE125 pKa = 4.43FISDD129 pKa = 4.46LPTAQPALEE138 pKa = 4.36SMEE141 pKa = 4.85SPLPPSDD148 pKa = 3.79LSEE151 pKa = 4.5FSWHH155 pKa = 5.4TSSRR159 pKa = 11.84LLEE162 pKa = 4.52SDD164 pKa = 4.95DD165 pKa = 3.8VPQASFTMSPGEE177 pKa = 4.02SWIEE181 pKa = 3.63EE182 pKa = 4.24QPFGEE187 pKa = 4.91HH188 pKa = 7.81DD189 pKa = 4.83DD190 pKa = 3.98DD191 pKa = 4.39TSDD194 pKa = 3.15GSAYY198 pKa = 10.38SLPGGRR204 pKa = 11.84THH206 pKa = 8.19PILFDD211 pKa = 3.33EE212 pKa = 5.09AEE214 pKa = 4.33WNGATWDD221 pKa = 4.08AVCLADD227 pKa = 4.59FLGGLFEE234 pKa = 6.02KK235 pKa = 10.63ADD237 pKa = 3.64PWSTLDD243 pKa = 4.98DD244 pKa = 4.37FLDD247 pKa = 4.23VPSLLPDD254 pKa = 4.31LPPTSTSLFLLASDD268 pKa = 4.18RR269 pKa = 11.84GGVGYY274 pKa = 10.52VATPRR279 pKa = 11.84SPDD282 pKa = 3.18WQDD285 pKa = 3.76LSSQTLAEE293 pKa = 4.3EE294 pKa = 4.43TRR296 pKa = 11.84ACTPLFSVSCGTEE309 pKa = 3.58ALEE312 pKa = 5.12DD313 pKa = 3.93VPAVDD318 pKa = 4.96SDD320 pKa = 4.21PPSSGDD326 pKa = 3.14TAYY329 pKa = 10.99DD330 pKa = 3.56DD331 pKa = 4.2TPEE334 pKa = 3.97EE335 pKa = 4.05AQAYY339 pKa = 8.93LPRR342 pKa = 11.84AEE344 pKa = 4.62YY345 pKa = 10.59EE346 pKa = 4.19PLSAPAMPSPSPVEE360 pKa = 4.03MLVKK364 pKa = 10.37PPGVTVRR371 pKa = 11.84GPCASIPAAEE381 pKa = 4.52EE382 pKa = 4.17DD383 pKa = 3.83SDD385 pKa = 4.67VLEE388 pKa = 4.81EE389 pKa = 4.32KK390 pKa = 10.4CGHH393 pKa = 6.59DD394 pKa = 4.32ASLGTDD400 pKa = 3.42GADD403 pKa = 2.92AAVEE407 pKa = 4.07RR408 pKa = 11.84GIEE411 pKa = 4.06TGVPEE416 pKa = 4.47SFDD419 pKa = 3.7RR420 pKa = 11.84DD421 pKa = 3.65VSVDD425 pKa = 3.69GPSLFADD432 pKa = 4.41DD433 pKa = 6.37DD434 pKa = 4.27VDD436 pKa = 6.16LDD438 pKa = 4.75DD439 pKa = 4.61EE440 pKa = 4.53
Molecular weight: 47.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A371DFU0|A0A371DFU0_9APHY Laccase C OS=Polyporus brumalis OX=139420 GN=OH76DRAFT_1417405 PE=3 SV=1
RR1 pKa = 7.38LHH3 pKa = 6.34KK4 pKa = 10.08QVPGLARR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.32KK14 pKa = 10.06QVPGLARR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.32KK24 pKa = 10.06QVPGLARR31 pKa = 11.84RR32 pKa = 11.84HH33 pKa = 5.32KK34 pKa = 10.06QVPGLARR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 5.32KK44 pKa = 10.06QVPGLARR51 pKa = 11.84RR52 pKa = 11.84HH53 pKa = 4.96KK54 pKa = 10.19QQ55 pKa = 2.94
RR1 pKa = 7.38LHH3 pKa = 6.34KK4 pKa = 10.08QVPGLARR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.32KK14 pKa = 10.06QVPGLARR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.32KK24 pKa = 10.06QVPGLARR31 pKa = 11.84RR32 pKa = 11.84HH33 pKa = 5.32KK34 pKa = 10.06QVPGLARR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 5.32KK44 pKa = 10.06QVPGLARR51 pKa = 11.84RR52 pKa = 11.84HH53 pKa = 4.96KK54 pKa = 10.19QQ55 pKa = 2.94
Molecular weight: 6.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7177947 |
49 |
5041 |
395.0 |
43.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.407 ± 0.018 | 1.459 ± 0.008 |
5.626 ± 0.013 | 5.732 ± 0.02 |
3.53 ± 0.01 | 6.517 ± 0.016 |
2.713 ± 0.009 | 4.252 ± 0.013 |
3.922 ± 0.017 | 9.242 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.06 ± 0.007 | 2.881 ± 0.009 |
6.946 ± 0.022 | 3.564 ± 0.013 |
7.028 ± 0.017 | 8.396 ± 0.023 |
5.973 ± 0.013 | 6.599 ± 0.015 |
1.512 ± 0.007 | 2.641 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
