
Leifsonia sp. Leaf336
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Leifsonia; unclassified Leifsonia
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
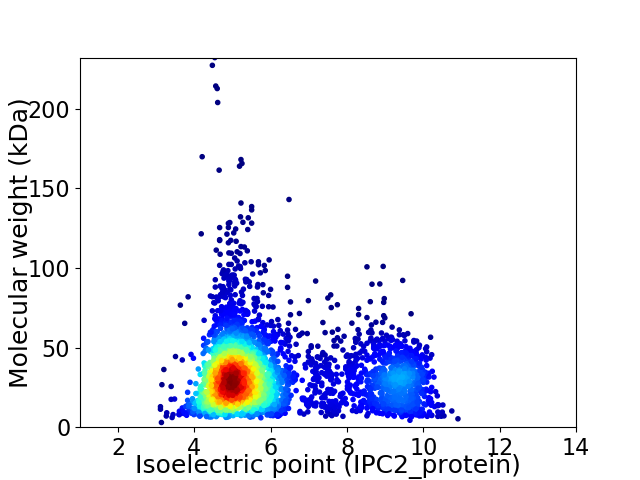
Virtual 2D-PAGE plot for 3857 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5MDL0|A0A0Q5MDL0_9MICO Hydroxyacid dehydrogenase OS=Leifsonia sp. Leaf336 OX=1736341 GN=ASF88_08080 PE=4 SV=1
MM1 pKa = 7.49LMSITAGAAKK11 pKa = 10.27RR12 pKa = 11.84STRR15 pKa = 11.84STLAFAAIIALLMTMLAVTSTNSSPADD42 pKa = 3.58AAQPWCGGTTVFGVVPDD59 pKa = 5.15DD60 pKa = 4.21PAASPTNTALVEE72 pKa = 4.07IDD74 pKa = 3.4GSSVGNAQLSGSLVTRR90 pKa = 11.84SPGDD94 pKa = 3.37TNLQNGLGLTADD106 pKa = 3.94GSTAYY111 pKa = 10.29AGADD115 pKa = 2.92NGTTIRR121 pKa = 11.84RR122 pKa = 11.84YY123 pKa = 11.27DD124 pKa = 3.73MLTDD128 pKa = 3.46TWTSYY133 pKa = 10.69PGRR136 pKa = 11.84PLPAGGEE143 pKa = 4.41GVVAGAVDD151 pKa = 4.31PVSGIYY157 pKa = 10.39YY158 pKa = 9.9FGEE161 pKa = 4.43IGAGPTDD168 pKa = 3.7PTLGHH173 pKa = 7.05LWGFDD178 pKa = 3.23TNTNQLISPAPLITFPSVAGEE199 pKa = 4.12NNGDD203 pKa = 3.49IAFDD207 pKa = 3.81GAGNLYY213 pKa = 9.98IVTSSTTHH221 pKa = 6.34GDD223 pKa = 3.69LEE225 pKa = 4.65VSPGPLPTTAAQAQLLQPTLLSGFTVPPAAGAAVNGIAFDD265 pKa = 3.8ATGTLYY271 pKa = 11.07LSLSGATGGVFSIDD285 pKa = 3.78PGTGAPQSALTPLSAALIGPSGTGLVDD312 pKa = 4.28LASCSFPPSVSAQKK326 pKa = 10.75NVLSRR331 pKa = 11.84VNPTDD336 pKa = 3.15QFTLSVTGPTVSLPATATTVGTADD360 pKa = 4.66GLQQTAGGQIVQAGPALARR379 pKa = 11.84FSDD382 pKa = 4.41TYY384 pKa = 10.66TATEE388 pKa = 4.25TASGTTDD395 pKa = 3.6LSQYY399 pKa = 8.65STTYY403 pKa = 10.16SCVDD407 pKa = 3.19VVNPTNPQFPITGSGTTVSFPLDD430 pKa = 3.61PFVGEE435 pKa = 4.47SPSVVCTFTNAALTSALTLEE455 pKa = 4.72KK456 pKa = 9.53TASPTAVSTVGQTVTYY472 pKa = 10.23SFLVTNTGSTILTSIAVAEE491 pKa = 4.38TAFSGTGTLPAPTCPTTTLDD511 pKa = 4.25PGDD514 pKa = 3.96STICTAAYY522 pKa = 9.96DD523 pKa = 3.69VTAADD528 pKa = 3.42IAAGVIDD535 pKa = 4.24NTAEE539 pKa = 4.1ANGLDD544 pKa = 3.96PSNDD548 pKa = 3.29TVTSPEE554 pKa = 4.19DD555 pKa = 3.51SAQVTATPAVAALDD569 pKa = 3.81LTKK572 pKa = 9.88TANPATVTAAGDD584 pKa = 3.74TITYY588 pKa = 7.38TFHH591 pKa = 5.54VTNTGNVPIDD601 pKa = 3.76SIAIQEE607 pKa = 4.52GNFSGTGSISAITCNATTLAPAASTDD633 pKa = 3.52CTATYY638 pKa = 9.16XITEE642 pKa = 4.26VVVLWHH648 pKa = 7.16RR649 pKa = 11.84DD650 pKa = 3.09RR651 pKa = 11.84GRR653 pKa = 3.89
MM1 pKa = 7.49LMSITAGAAKK11 pKa = 10.27RR12 pKa = 11.84STRR15 pKa = 11.84STLAFAAIIALLMTMLAVTSTNSSPADD42 pKa = 3.58AAQPWCGGTTVFGVVPDD59 pKa = 5.15DD60 pKa = 4.21PAASPTNTALVEE72 pKa = 4.07IDD74 pKa = 3.4GSSVGNAQLSGSLVTRR90 pKa = 11.84SPGDD94 pKa = 3.37TNLQNGLGLTADD106 pKa = 3.94GSTAYY111 pKa = 10.29AGADD115 pKa = 2.92NGTTIRR121 pKa = 11.84RR122 pKa = 11.84YY123 pKa = 11.27DD124 pKa = 3.73MLTDD128 pKa = 3.46TWTSYY133 pKa = 10.69PGRR136 pKa = 11.84PLPAGGEE143 pKa = 4.41GVVAGAVDD151 pKa = 4.31PVSGIYY157 pKa = 10.39YY158 pKa = 9.9FGEE161 pKa = 4.43IGAGPTDD168 pKa = 3.7PTLGHH173 pKa = 7.05LWGFDD178 pKa = 3.23TNTNQLISPAPLITFPSVAGEE199 pKa = 4.12NNGDD203 pKa = 3.49IAFDD207 pKa = 3.81GAGNLYY213 pKa = 9.98IVTSSTTHH221 pKa = 6.34GDD223 pKa = 3.69LEE225 pKa = 4.65VSPGPLPTTAAQAQLLQPTLLSGFTVPPAAGAAVNGIAFDD265 pKa = 3.8ATGTLYY271 pKa = 11.07LSLSGATGGVFSIDD285 pKa = 3.78PGTGAPQSALTPLSAALIGPSGTGLVDD312 pKa = 4.28LASCSFPPSVSAQKK326 pKa = 10.75NVLSRR331 pKa = 11.84VNPTDD336 pKa = 3.15QFTLSVTGPTVSLPATATTVGTADD360 pKa = 4.66GLQQTAGGQIVQAGPALARR379 pKa = 11.84FSDD382 pKa = 4.41TYY384 pKa = 10.66TATEE388 pKa = 4.25TASGTTDD395 pKa = 3.6LSQYY399 pKa = 8.65STTYY403 pKa = 10.16SCVDD407 pKa = 3.19VVNPTNPQFPITGSGTTVSFPLDD430 pKa = 3.61PFVGEE435 pKa = 4.47SPSVVCTFTNAALTSALTLEE455 pKa = 4.72KK456 pKa = 9.53TASPTAVSTVGQTVTYY472 pKa = 10.23SFLVTNTGSTILTSIAVAEE491 pKa = 4.38TAFSGTGTLPAPTCPTTTLDD511 pKa = 4.25PGDD514 pKa = 3.96STICTAAYY522 pKa = 9.96DD523 pKa = 3.69VTAADD528 pKa = 3.42IAAGVIDD535 pKa = 4.24NTAEE539 pKa = 4.1ANGLDD544 pKa = 3.96PSNDD548 pKa = 3.29TVTSPEE554 pKa = 4.19DD555 pKa = 3.51SAQVTATPAVAALDD569 pKa = 3.81LTKK572 pKa = 9.88TANPATVTAAGDD584 pKa = 3.74TITYY588 pKa = 7.38TFHH591 pKa = 5.54VTNTGNVPIDD601 pKa = 3.76SIAIQEE607 pKa = 4.52GNFSGTGSISAITCNATTLAPAASTDD633 pKa = 3.52CTATYY638 pKa = 9.16XITEE642 pKa = 4.26VVVLWHH648 pKa = 7.16RR649 pKa = 11.84DD650 pKa = 3.09RR651 pKa = 11.84GRR653 pKa = 3.89
Molecular weight: 65.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5M3B3|A0A0Q5M3B3_9MICO Phosphoglucomutase OS=Leifsonia sp. Leaf336 OX=1736341 GN=ASF88_16840 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.52GRR40 pKa = 11.84EE41 pKa = 3.9KK42 pKa = 10.95LSAA45 pKa = 3.78
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.52GRR40 pKa = 11.84EE41 pKa = 3.9KK42 pKa = 10.95LSAA45 pKa = 3.78
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1226354 |
27 |
2233 |
318.0 |
33.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.502 ± 0.056 | 0.472 ± 0.01 |
6.041 ± 0.033 | 5.421 ± 0.047 |
3.26 ± 0.023 | 9.029 ± 0.037 |
1.933 ± 0.016 | 4.445 ± 0.03 |
2.035 ± 0.028 | 10.261 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.648 ± 0.014 | 2.088 ± 0.025 |
5.41 ± 0.027 | 2.816 ± 0.023 |
7.072 ± 0.048 | 5.793 ± 0.029 |
6.167 ± 0.041 | 8.987 ± 0.038 |
1.524 ± 0.019 | 2.095 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
