
Labilithrix luteola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Sorangiineae; Labilitrichaceae; Labilithrix
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
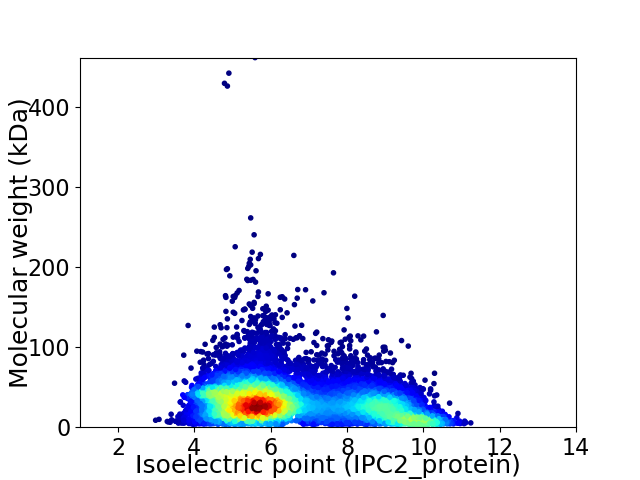
Virtual 2D-PAGE plot for 11481 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1PLF3|A0A0K1PLF3_9DELT Response regulator of zinc sigma-54-dependent two-component system OS=Labilithrix luteola OX=1391654 GN=AKJ09_00613 PE=4 SV=1
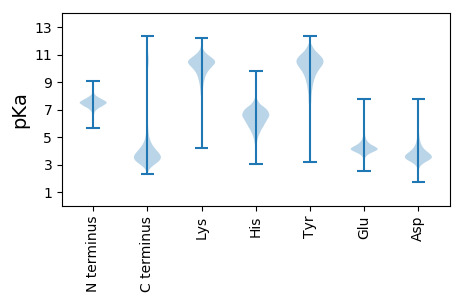
MM1 pKa = 7.39QGRR4 pKa = 11.84KK5 pKa = 9.56DD6 pKa = 3.63GQEE9 pKa = 3.87GGEE12 pKa = 4.51TNGSPGTACPTDD24 pKa = 3.36AGGADD29 pKa = 3.54TSLPPGDD36 pKa = 4.63AQPDD40 pKa = 3.5AHH42 pKa = 7.1VDD44 pKa = 3.48PDD46 pKa = 4.62GPPLDD51 pKa = 4.79ASTDD55 pKa = 3.77DD56 pKa = 4.88VVDD59 pKa = 4.66ASCPGDD65 pKa = 3.97DD66 pKa = 4.01AAADD70 pKa = 3.84SGMDD74 pKa = 3.51AADD77 pKa = 3.66ATTDD81 pKa = 3.6ASDD84 pKa = 3.36AAQPEE89 pKa = 4.57DD90 pKa = 3.88GGSDD94 pKa = 3.4ASDD97 pKa = 4.17GGPSDD102 pKa = 4.13AGSDD106 pKa = 3.76VNVPPDD112 pKa = 3.28ASSDD116 pKa = 3.76VTCTFTRR123 pKa = 11.84SHH125 pKa = 6.43TGANDD130 pKa = 3.18ACNYY134 pKa = 9.54CVLPPEE140 pKa = 4.08VTISIEE146 pKa = 4.77HH147 pKa = 5.35VPLWIDD153 pKa = 3.23HH154 pKa = 6.44GEE156 pKa = 4.05HH157 pKa = 6.11GVGIEE162 pKa = 4.3GQGDD166 pKa = 4.74LPPQPPPPVPPKK178 pKa = 10.47RR179 pKa = 11.84VIHH182 pKa = 6.01VDD184 pKa = 2.95APTVFDD190 pKa = 3.85SWWMFGDD197 pKa = 3.91AFVLSGTPPSEE208 pKa = 4.54SYY210 pKa = 11.03VSSNMPFVPNGGFNGTITVGSGNATSLTLNGSHH243 pKa = 7.2FPATFGCVALHH254 pKa = 6.04GCFVEE259 pKa = 6.33KK260 pKa = 10.26IACTGTGIAEE270 pKa = 4.08NRR272 pKa = 3.62
MM1 pKa = 7.39QGRR4 pKa = 11.84KK5 pKa = 9.56DD6 pKa = 3.63GQEE9 pKa = 3.87GGEE12 pKa = 4.51TNGSPGTACPTDD24 pKa = 3.36AGGADD29 pKa = 3.54TSLPPGDD36 pKa = 4.63AQPDD40 pKa = 3.5AHH42 pKa = 7.1VDD44 pKa = 3.48PDD46 pKa = 4.62GPPLDD51 pKa = 4.79ASTDD55 pKa = 3.77DD56 pKa = 4.88VVDD59 pKa = 4.66ASCPGDD65 pKa = 3.97DD66 pKa = 4.01AAADD70 pKa = 3.84SGMDD74 pKa = 3.51AADD77 pKa = 3.66ATTDD81 pKa = 3.6ASDD84 pKa = 3.36AAQPEE89 pKa = 4.57DD90 pKa = 3.88GGSDD94 pKa = 3.4ASDD97 pKa = 4.17GGPSDD102 pKa = 4.13AGSDD106 pKa = 3.76VNVPPDD112 pKa = 3.28ASSDD116 pKa = 3.76VTCTFTRR123 pKa = 11.84SHH125 pKa = 6.43TGANDD130 pKa = 3.18ACNYY134 pKa = 9.54CVLPPEE140 pKa = 4.08VTISIEE146 pKa = 4.77HH147 pKa = 5.35VPLWIDD153 pKa = 3.23HH154 pKa = 6.44GEE156 pKa = 4.05HH157 pKa = 6.11GVGIEE162 pKa = 4.3GQGDD166 pKa = 4.74LPPQPPPPVPPKK178 pKa = 10.47RR179 pKa = 11.84VIHH182 pKa = 6.01VDD184 pKa = 2.95APTVFDD190 pKa = 3.85SWWMFGDD197 pKa = 3.91AFVLSGTPPSEE208 pKa = 4.54SYY210 pKa = 11.03VSSNMPFVPNGGFNGTITVGSGNATSLTLNGSHH243 pKa = 7.2FPATFGCVALHH254 pKa = 6.04GCFVEE259 pKa = 6.33KK260 pKa = 10.26IACTGTGIAEE270 pKa = 4.08NRR272 pKa = 3.62
Molecular weight: 27.39 kDa
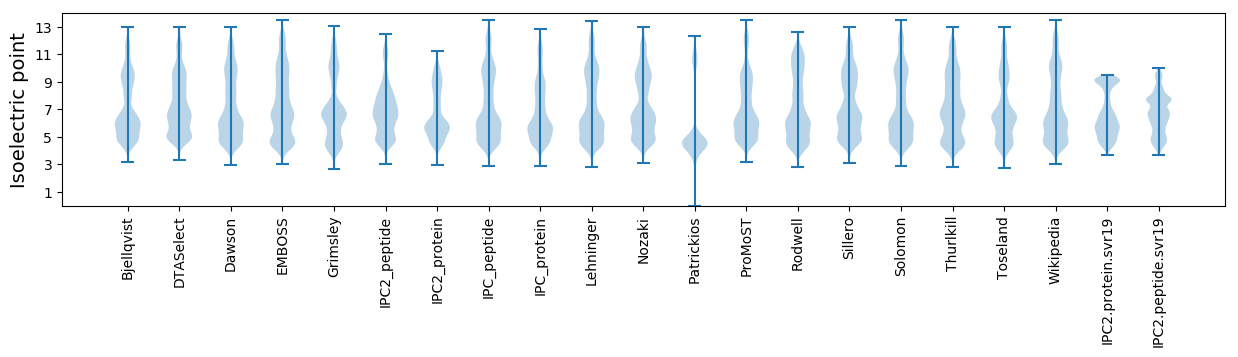
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1Q8I9|A0A0K1Q8I9_9DELT Uncharacterized protein OS=Labilithrix luteola OX=1391654 GN=AKJ09_08789 PE=4 SV=1
MM1 pKa = 7.55LLGSSRR7 pKa = 11.84LRR9 pKa = 11.84LRR11 pKa = 11.84KK12 pKa = 8.79RR13 pKa = 11.84TSLHH17 pKa = 5.34ARR19 pKa = 11.84AAFAARR25 pKa = 11.84RR26 pKa = 11.84VARR29 pKa = 11.84GSRR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 5.31QPANKK39 pKa = 8.76HH40 pKa = 5.66KK41 pKa = 10.54KK42 pKa = 8.14GTTIRR47 pKa = 3.67
MM1 pKa = 7.55LLGSSRR7 pKa = 11.84LRR9 pKa = 11.84LRR11 pKa = 11.84KK12 pKa = 8.79RR13 pKa = 11.84TSLHH17 pKa = 5.34ARR19 pKa = 11.84AAFAARR25 pKa = 11.84RR26 pKa = 11.84VARR29 pKa = 11.84GSRR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 5.31QPANKK39 pKa = 8.76HH40 pKa = 5.66KK41 pKa = 10.54KK42 pKa = 8.14GTTIRR47 pKa = 3.67
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3621879 |
37 |
4320 |
315.5 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.638 ± 0.032 | 1.324 ± 0.022 |
5.878 ± 0.02 | 5.76 ± 0.028 |
3.357 ± 0.016 | 8.683 ± 0.027 |
2.016 ± 0.01 | 3.783 ± 0.013 |
3.294 ± 0.018 | 9.276 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.01 | 2.387 ± 0.014 |
5.894 ± 0.022 | 2.585 ± 0.012 |
7.5 ± 0.029 | 6.429 ± 0.02 |
5.927 ± 0.021 | 8.146 ± 0.02 |
1.279 ± 0.009 | 1.946 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
