
Wenyingzhuangia fucanilytica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Wenyingzhuangia
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
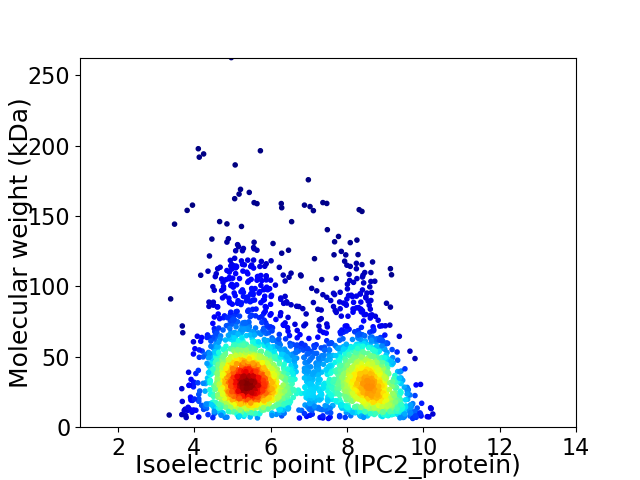
Virtual 2D-PAGE plot for 2747 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B1Y4Y1|A0A1B1Y4Y1_9FLAO Cyclic nucleotide-binding protein OS=Wenyingzhuangia fucanilytica OX=1790137 GN=AXE80_05665 PE=4 SV=1
MM1 pKa = 7.26KK2 pKa = 9.52TLKK5 pKa = 10.74LITALMFVSFLGFISCQKK23 pKa = 10.67EE24 pKa = 3.42IDD26 pKa = 4.01EE27 pKa = 5.12EE28 pKa = 4.25NGQNPNTNSVNSEE41 pKa = 3.94TAKK44 pKa = 10.62NLQRR48 pKa = 11.84TAMYY52 pKa = 10.27DD53 pKa = 3.3GSFDD57 pKa = 4.29DD58 pKa = 5.88FLDD61 pKa = 4.22GTSCSSVLFPVTVWVNNQKK80 pKa = 10.49IILTNKK86 pKa = 9.8NDD88 pKa = 3.54YY89 pKa = 10.86QLVLNILGEE98 pKa = 4.35VINDD102 pKa = 3.16EE103 pKa = 4.19DD104 pKa = 4.61TITFQFPISVKK115 pKa = 9.92LSNYY119 pKa = 9.4TEE121 pKa = 4.15VQVEE125 pKa = 4.03NQAEE129 pKa = 4.16YY130 pKa = 11.16NAIKK134 pKa = 9.05DD135 pKa = 3.62TCEE138 pKa = 3.32EE139 pKa = 4.09AEE141 pKa = 4.2AEE143 pKa = 4.15KK144 pKa = 11.03EE145 pKa = 4.07NAINCLKK152 pKa = 9.94MNYY155 pKa = 8.86PISILTYY162 pKa = 10.33NISLEE167 pKa = 4.27QTSTTVIEE175 pKa = 4.78SNQEE179 pKa = 3.63LYY181 pKa = 10.63TYY183 pKa = 9.73MSNFNEE189 pKa = 4.36DD190 pKa = 3.04EE191 pKa = 4.3KK192 pKa = 11.42FSIKK196 pKa = 10.68YY197 pKa = 8.91PISATLSNDD206 pKa = 3.25TEE208 pKa = 4.57VNISSDD214 pKa = 3.9AEE216 pKa = 4.34FKK218 pKa = 11.05SHH220 pKa = 7.49ISDD223 pKa = 3.88CLGDD227 pKa = 4.48EE228 pKa = 4.36DD229 pKa = 5.45TMDD232 pKa = 4.91DD233 pKa = 3.73AQEE236 pKa = 4.18DD237 pKa = 4.57AEE239 pKa = 4.31TLEE242 pKa = 4.66NILVEE247 pKa = 4.17GTFRR251 pKa = 11.84VQSYY255 pKa = 10.98INGGVDD261 pKa = 3.35TANTYY266 pKa = 11.37ANYY269 pKa = 9.83TIDD272 pKa = 3.57FANDD276 pKa = 3.66LTCSATNTANALAEE290 pKa = 4.22KK291 pKa = 10.9AEE293 pKa = 4.42GTFSVTSEE301 pKa = 4.02MEE303 pKa = 3.92VFLTLEE309 pKa = 4.42FSSEE313 pKa = 4.18ANFEE317 pKa = 4.46LLNDD321 pKa = 3.35TWTVTNYY328 pKa = 10.09SSSSITLQSTTNTSTALVLNQII350 pKa = 4.41
MM1 pKa = 7.26KK2 pKa = 9.52TLKK5 pKa = 10.74LITALMFVSFLGFISCQKK23 pKa = 10.67EE24 pKa = 3.42IDD26 pKa = 4.01EE27 pKa = 5.12EE28 pKa = 4.25NGQNPNTNSVNSEE41 pKa = 3.94TAKK44 pKa = 10.62NLQRR48 pKa = 11.84TAMYY52 pKa = 10.27DD53 pKa = 3.3GSFDD57 pKa = 4.29DD58 pKa = 5.88FLDD61 pKa = 4.22GTSCSSVLFPVTVWVNNQKK80 pKa = 10.49IILTNKK86 pKa = 9.8NDD88 pKa = 3.54YY89 pKa = 10.86QLVLNILGEE98 pKa = 4.35VINDD102 pKa = 3.16EE103 pKa = 4.19DD104 pKa = 4.61TITFQFPISVKK115 pKa = 9.92LSNYY119 pKa = 9.4TEE121 pKa = 4.15VQVEE125 pKa = 4.03NQAEE129 pKa = 4.16YY130 pKa = 11.16NAIKK134 pKa = 9.05DD135 pKa = 3.62TCEE138 pKa = 3.32EE139 pKa = 4.09AEE141 pKa = 4.2AEE143 pKa = 4.15KK144 pKa = 11.03EE145 pKa = 4.07NAINCLKK152 pKa = 9.94MNYY155 pKa = 8.86PISILTYY162 pKa = 10.33NISLEE167 pKa = 4.27QTSTTVIEE175 pKa = 4.78SNQEE179 pKa = 3.63LYY181 pKa = 10.63TYY183 pKa = 9.73MSNFNEE189 pKa = 4.36DD190 pKa = 3.04EE191 pKa = 4.3KK192 pKa = 11.42FSIKK196 pKa = 10.68YY197 pKa = 8.91PISATLSNDD206 pKa = 3.25TEE208 pKa = 4.57VNISSDD214 pKa = 3.9AEE216 pKa = 4.34FKK218 pKa = 11.05SHH220 pKa = 7.49ISDD223 pKa = 3.88CLGDD227 pKa = 4.48EE228 pKa = 4.36DD229 pKa = 5.45TMDD232 pKa = 4.91DD233 pKa = 3.73AQEE236 pKa = 4.18DD237 pKa = 4.57AEE239 pKa = 4.31TLEE242 pKa = 4.66NILVEE247 pKa = 4.17GTFRR251 pKa = 11.84VQSYY255 pKa = 10.98INGGVDD261 pKa = 3.35TANTYY266 pKa = 11.37ANYY269 pKa = 9.83TIDD272 pKa = 3.57FANDD276 pKa = 3.66LTCSATNTANALAEE290 pKa = 4.22KK291 pKa = 10.9AEE293 pKa = 4.42GTFSVTSEE301 pKa = 4.02MEE303 pKa = 3.92VFLTLEE309 pKa = 4.42FSSEE313 pKa = 4.18ANFEE317 pKa = 4.46LLNDD321 pKa = 3.35TWTVTNYY328 pKa = 10.09SSSSITLQSTTNTSTALVLNQII350 pKa = 4.41
Molecular weight: 38.99 kDa
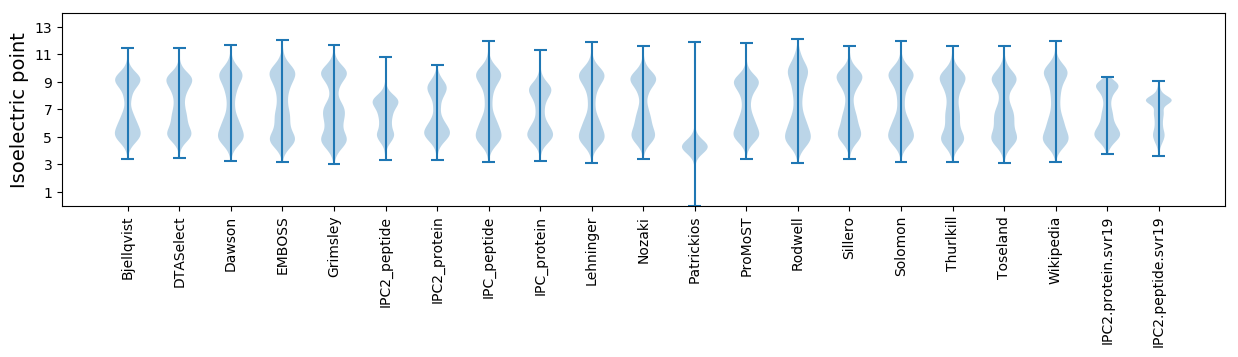
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B1Y7W9|A0A1B1Y7W9_9FLAO 30S ribosomal protein S12 OS=Wenyingzhuangia fucanilytica OX=1790137 GN=rpsL PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.58SLLAPNKK38 pKa = 10.19RR39 pKa = 11.84SGGRR43 pKa = 11.84NNSGRR48 pKa = 11.84MTMRR52 pKa = 11.84YY53 pKa = 9.71VGGGHH58 pKa = 6.68KK59 pKa = 9.72KK60 pKa = 9.76RR61 pKa = 11.84YY62 pKa = 9.53RR63 pKa = 11.84IIDD66 pKa = 3.52FNRR69 pKa = 11.84DD70 pKa = 3.14KK71 pKa = 11.15FGVEE75 pKa = 3.76ATVKK79 pKa = 9.91TIEE82 pKa = 4.03YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALVEE95 pKa = 4.25YY96 pKa = 10.5TDD98 pKa = 3.61GEE100 pKa = 4.17KK101 pKa = 10.32RR102 pKa = 11.84YY103 pKa = 10.63VIAQAGLQVGQTIVSNNDD121 pKa = 3.32AITPDD126 pKa = 3.47VGNAMPLANIPLGTTICCIEE146 pKa = 4.14LRR148 pKa = 11.84PGQGAVMARR157 pKa = 11.84SAGSFAQLMAKK168 pKa = 8.96EE169 pKa = 4.42GKK171 pKa = 9.47YY172 pKa = 9.15ATVKK176 pKa = 10.52LPSGEE181 pKa = 4.06TRR183 pKa = 11.84LILLTCIATIGVVSNSDD200 pKa = 3.21HH201 pKa = 6.09QLLVSGKK208 pKa = 9.49AGRR211 pKa = 11.84SRR213 pKa = 11.84WLGRR217 pKa = 11.84RR218 pKa = 11.84PRR220 pKa = 11.84TNAVRR225 pKa = 11.84MNPVDD230 pKa = 3.4HH231 pKa = 7.07PMGGGEE237 pKa = 4.08GRR239 pKa = 11.84SSGGHH244 pKa = 4.7PRR246 pKa = 11.84SRR248 pKa = 11.84NGIPAKK254 pKa = 10.28GYY256 pKa = 7.21KK257 pKa = 8.8TRR259 pKa = 11.84SKK261 pKa = 9.63TKK263 pKa = 10.38ASNKK267 pKa = 9.94YY268 pKa = 8.99ILEE271 pKa = 4.09RR272 pKa = 11.84RR273 pKa = 11.84KK274 pKa = 10.2KK275 pKa = 9.94
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.58SLLAPNKK38 pKa = 10.19RR39 pKa = 11.84SGGRR43 pKa = 11.84NNSGRR48 pKa = 11.84MTMRR52 pKa = 11.84YY53 pKa = 9.71VGGGHH58 pKa = 6.68KK59 pKa = 9.72KK60 pKa = 9.76RR61 pKa = 11.84YY62 pKa = 9.53RR63 pKa = 11.84IIDD66 pKa = 3.52FNRR69 pKa = 11.84DD70 pKa = 3.14KK71 pKa = 11.15FGVEE75 pKa = 3.76ATVKK79 pKa = 9.91TIEE82 pKa = 4.03YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALVEE95 pKa = 4.25YY96 pKa = 10.5TDD98 pKa = 3.61GEE100 pKa = 4.17KK101 pKa = 10.32RR102 pKa = 11.84YY103 pKa = 10.63VIAQAGLQVGQTIVSNNDD121 pKa = 3.32AITPDD126 pKa = 3.47VGNAMPLANIPLGTTICCIEE146 pKa = 4.14LRR148 pKa = 11.84PGQGAVMARR157 pKa = 11.84SAGSFAQLMAKK168 pKa = 8.96EE169 pKa = 4.42GKK171 pKa = 9.47YY172 pKa = 9.15ATVKK176 pKa = 10.52LPSGEE181 pKa = 4.06TRR183 pKa = 11.84LILLTCIATIGVVSNSDD200 pKa = 3.21HH201 pKa = 6.09QLLVSGKK208 pKa = 9.49AGRR211 pKa = 11.84SRR213 pKa = 11.84WLGRR217 pKa = 11.84RR218 pKa = 11.84PRR220 pKa = 11.84TNAVRR225 pKa = 11.84MNPVDD230 pKa = 3.4HH231 pKa = 7.07PMGGGEE237 pKa = 4.08GRR239 pKa = 11.84SSGGHH244 pKa = 4.7PRR246 pKa = 11.84SRR248 pKa = 11.84NGIPAKK254 pKa = 10.28GYY256 pKa = 7.21KK257 pKa = 8.8TRR259 pKa = 11.84SKK261 pKa = 9.63TKK263 pKa = 10.38ASNKK267 pKa = 9.94YY268 pKa = 8.99ILEE271 pKa = 4.09RR272 pKa = 11.84RR273 pKa = 11.84KK274 pKa = 10.2KK275 pKa = 9.94
Molecular weight: 30.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1031829 |
54 |
2341 |
375.6 |
42.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.177 ± 0.043 | 0.756 ± 0.011 |
5.519 ± 0.032 | 6.486 ± 0.04 |
5.026 ± 0.03 | 6.461 ± 0.041 |
1.876 ± 0.019 | 7.883 ± 0.046 |
8.25 ± 0.05 | 9.008 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.103 ± 0.019 | 6.597 ± 0.05 |
3.397 ± 0.023 | 3.353 ± 0.021 |
3.066 ± 0.025 | 6.332 ± 0.038 |
5.983 ± 0.04 | 6.394 ± 0.037 |
1.115 ± 0.02 | 4.218 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
