
Capybara microvirus Cap3_SP_393
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
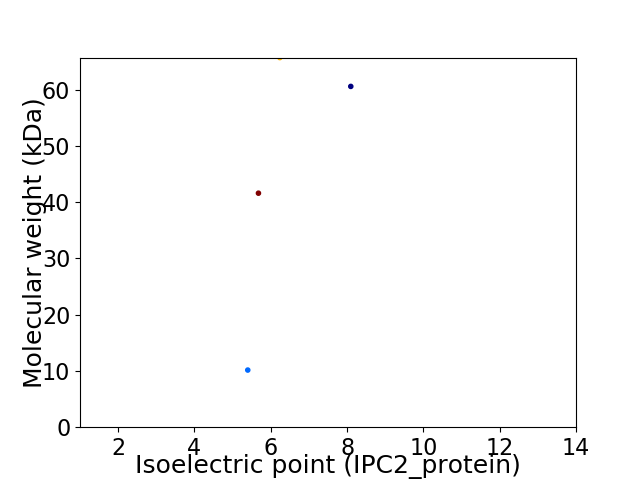
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVS9|A0A4V1FVS9_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_393 OX=2585443 PE=4 SV=1
MM1 pKa = 6.99STEE4 pKa = 4.16KK5 pKa = 10.33EE6 pKa = 3.97QKK8 pKa = 10.17KK9 pKa = 10.06KK10 pKa = 10.56FFRR13 pKa = 11.84SLIEE17 pKa = 3.83RR18 pKa = 11.84TKK20 pKa = 10.82TKK22 pKa = 10.54LPPKK26 pKa = 9.75AVSVVLGFNNVVTLQTDD43 pKa = 4.11CPNPEE48 pKa = 3.92PDD50 pKa = 4.47FRR52 pKa = 11.84DD53 pKa = 3.23ISLGAQAEE61 pKa = 4.55SGSFLGEE68 pKa = 3.86VNTNNGDD75 pKa = 2.85IFTAHH80 pKa = 7.41KK81 pKa = 10.27LAEE84 pKa = 4.41DD85 pKa = 3.66VLNNSDD91 pKa = 3.23NN92 pKa = 3.59
MM1 pKa = 6.99STEE4 pKa = 4.16KK5 pKa = 10.33EE6 pKa = 3.97QKK8 pKa = 10.17KK9 pKa = 10.06KK10 pKa = 10.56FFRR13 pKa = 11.84SLIEE17 pKa = 3.83RR18 pKa = 11.84TKK20 pKa = 10.82TKK22 pKa = 10.54LPPKK26 pKa = 9.75AVSVVLGFNNVVTLQTDD43 pKa = 4.11CPNPEE48 pKa = 3.92PDD50 pKa = 4.47FRR52 pKa = 11.84DD53 pKa = 3.23ISLGAQAEE61 pKa = 4.55SGSFLGEE68 pKa = 3.86VNTNNGDD75 pKa = 2.85IFTAHH80 pKa = 7.41KK81 pKa = 10.27LAEE84 pKa = 4.41DD85 pKa = 3.66VLNNSDD91 pKa = 3.23NN92 pKa = 3.59
Molecular weight: 10.15 kDa
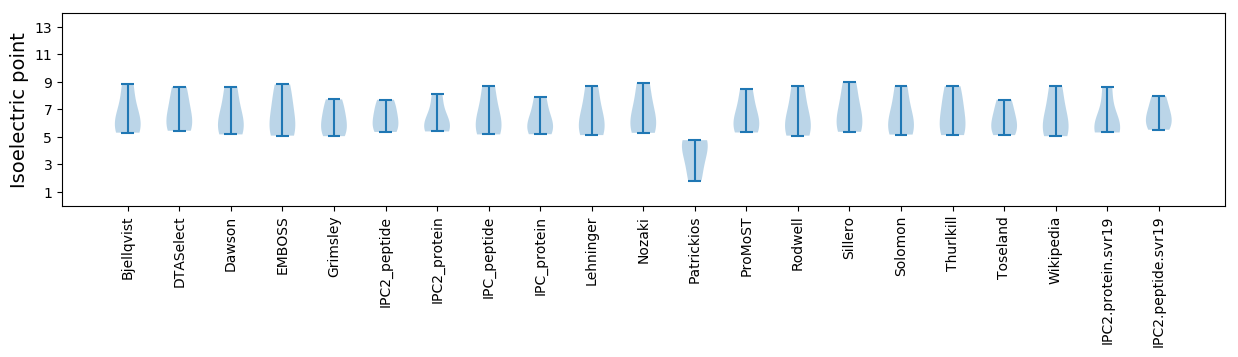
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8G9|A0A4P8W8G9_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_393 OX=2585443 PE=3 SV=1
MM1 pKa = 7.77LIKK4 pKa = 10.82NNSKK8 pKa = 9.24TSYY11 pKa = 9.85CSKK14 pKa = 10.23KK15 pKa = 10.5QYY17 pKa = 11.14ININDD22 pKa = 3.74SYY24 pKa = 11.53NYY26 pKa = 10.13LSSFQAYY33 pKa = 7.99VRR35 pKa = 11.84TYY37 pKa = 10.33YY38 pKa = 10.59DD39 pKa = 3.29YY40 pKa = 11.47QYY42 pKa = 10.71VQSNYY47 pKa = 9.26GVAYY51 pKa = 9.63FYY53 pKa = 10.87TLTFNDD59 pKa = 4.28LNLNTINYY67 pKa = 9.52NGISINTHH75 pKa = 3.56NHH77 pKa = 5.83KK78 pKa = 10.42LLHH81 pKa = 5.75LWIDD85 pKa = 3.51NVYY88 pKa = 10.63KK89 pKa = 10.64ILLRR93 pKa = 11.84KK94 pKa = 9.41FDD96 pKa = 3.86CKK98 pKa = 10.99SRR100 pKa = 11.84FFITSEE106 pKa = 3.93CGEE109 pKa = 4.37GKK111 pKa = 10.57GSRR114 pKa = 11.84KK115 pKa = 9.74KK116 pKa = 11.01GNNPHH121 pKa = 5.67YY122 pKa = 10.51HH123 pKa = 6.04VILYY127 pKa = 7.22VIPTIEE133 pKa = 4.22NMLPTSKK140 pKa = 10.72EE141 pKa = 3.56MLSIVRR147 pKa = 11.84YY148 pKa = 8.58CWQGTDD154 pKa = 4.47LDD156 pKa = 4.31LTLKK160 pKa = 9.84QNSYY164 pKa = 9.84IAKK167 pKa = 7.37TCKK170 pKa = 10.29NGILKK175 pKa = 10.14EE176 pKa = 4.09GKK178 pKa = 9.38NLGLIEE184 pKa = 4.31SLNALSYY191 pKa = 8.23VCKK194 pKa = 10.45YY195 pKa = 9.33VSKK198 pKa = 10.68DD199 pKa = 3.03IEE201 pKa = 4.57TKK203 pKa = 8.8EE204 pKa = 3.95HH205 pKa = 6.25YY206 pKa = 10.44DD207 pKa = 3.35IIKK210 pKa = 10.21NYY212 pKa = 10.5YY213 pKa = 8.0YY214 pKa = 11.13YY215 pKa = 11.21KK216 pKa = 10.58FYY218 pKa = 11.18NEE220 pKa = 4.7KK221 pKa = 10.57LLSCLKK227 pKa = 10.8CIDD230 pKa = 3.56NLIYY234 pKa = 10.36ILQKK238 pKa = 10.54SGLNEE243 pKa = 4.24TYY245 pKa = 10.66CNEE248 pKa = 3.88IQLDD252 pKa = 4.2NINSIDD258 pKa = 3.85EE259 pKa = 4.34FEE261 pKa = 5.44LYY263 pKa = 10.99LFDD266 pKa = 4.9FVKK269 pKa = 10.78NNHH272 pKa = 6.12GSTEE276 pKa = 4.48FILCNINKK284 pKa = 9.89LYY286 pKa = 10.79KK287 pKa = 10.28DD288 pKa = 3.16IYY290 pKa = 7.77NTYY293 pKa = 10.13RR294 pKa = 11.84LIHH297 pKa = 6.45KK298 pKa = 9.79EE299 pKa = 3.88YY300 pKa = 11.01DD301 pKa = 3.26FLDD304 pKa = 4.39YY305 pKa = 10.52IPKK308 pKa = 8.96TFYY311 pKa = 10.17PQMMGFKK318 pKa = 9.85LAKK321 pKa = 9.21QNIVEE326 pKa = 4.48TCSGTFIKK334 pKa = 9.83IDD336 pKa = 3.46NKK338 pKa = 10.43LCPISKK344 pKa = 10.26CFFNKK349 pKa = 10.32LYY351 pKa = 8.8KK352 pKa = 9.28THH354 pKa = 6.79LRR356 pKa = 11.84SCPSYY361 pKa = 11.09SRR363 pKa = 11.84YY364 pKa = 10.58NYY366 pKa = 10.68DD367 pKa = 3.93GINYY371 pKa = 9.36LCRR374 pKa = 11.84TIQNRR379 pKa = 11.84IDD381 pKa = 3.25ILAAKK386 pKa = 9.97ANLYY390 pKa = 11.09CNDD393 pKa = 3.33KK394 pKa = 10.7NICKK398 pKa = 9.99LYY400 pKa = 10.94SMYY403 pKa = 11.12KK404 pKa = 9.88LVYY407 pKa = 7.55EE408 pKa = 4.53HH409 pKa = 7.29RR410 pKa = 11.84LIPISDD416 pKa = 3.7FNKK419 pKa = 9.44PLEE422 pKa = 3.95MLKK425 pKa = 10.38DD426 pKa = 3.58YY427 pKa = 10.95KK428 pKa = 11.21NYY430 pKa = 9.89MEE432 pKa = 5.15LEE434 pKa = 4.47SIFSFRR440 pKa = 11.84RR441 pKa = 11.84TFGIEE446 pKa = 4.01LNLDD450 pKa = 3.61NYY452 pKa = 9.9EE453 pKa = 4.08KK454 pKa = 10.87AIMHH458 pKa = 5.98PQFYY462 pKa = 9.35PYY464 pKa = 10.52RR465 pKa = 11.84SIFTSISNVIEE476 pKa = 4.06KK477 pKa = 10.68SEE479 pKa = 4.24IADD482 pKa = 3.64KK483 pKa = 9.27QHH485 pKa = 6.76KK486 pKa = 9.64EE487 pKa = 3.71EE488 pKa = 4.09NFEE491 pKa = 4.27HH492 pKa = 6.62KK493 pKa = 10.58KK494 pKa = 10.05HH495 pKa = 5.69VKK497 pKa = 10.03KK498 pKa = 10.77LLNSYY503 pKa = 9.8RR504 pKa = 11.84YY505 pKa = 9.44EE506 pKa = 4.14YY507 pKa = 11.42
MM1 pKa = 7.77LIKK4 pKa = 10.82NNSKK8 pKa = 9.24TSYY11 pKa = 9.85CSKK14 pKa = 10.23KK15 pKa = 10.5QYY17 pKa = 11.14ININDD22 pKa = 3.74SYY24 pKa = 11.53NYY26 pKa = 10.13LSSFQAYY33 pKa = 7.99VRR35 pKa = 11.84TYY37 pKa = 10.33YY38 pKa = 10.59DD39 pKa = 3.29YY40 pKa = 11.47QYY42 pKa = 10.71VQSNYY47 pKa = 9.26GVAYY51 pKa = 9.63FYY53 pKa = 10.87TLTFNDD59 pKa = 4.28LNLNTINYY67 pKa = 9.52NGISINTHH75 pKa = 3.56NHH77 pKa = 5.83KK78 pKa = 10.42LLHH81 pKa = 5.75LWIDD85 pKa = 3.51NVYY88 pKa = 10.63KK89 pKa = 10.64ILLRR93 pKa = 11.84KK94 pKa = 9.41FDD96 pKa = 3.86CKK98 pKa = 10.99SRR100 pKa = 11.84FFITSEE106 pKa = 3.93CGEE109 pKa = 4.37GKK111 pKa = 10.57GSRR114 pKa = 11.84KK115 pKa = 9.74KK116 pKa = 11.01GNNPHH121 pKa = 5.67YY122 pKa = 10.51HH123 pKa = 6.04VILYY127 pKa = 7.22VIPTIEE133 pKa = 4.22NMLPTSKK140 pKa = 10.72EE141 pKa = 3.56MLSIVRR147 pKa = 11.84YY148 pKa = 8.58CWQGTDD154 pKa = 4.47LDD156 pKa = 4.31LTLKK160 pKa = 9.84QNSYY164 pKa = 9.84IAKK167 pKa = 7.37TCKK170 pKa = 10.29NGILKK175 pKa = 10.14EE176 pKa = 4.09GKK178 pKa = 9.38NLGLIEE184 pKa = 4.31SLNALSYY191 pKa = 8.23VCKK194 pKa = 10.45YY195 pKa = 9.33VSKK198 pKa = 10.68DD199 pKa = 3.03IEE201 pKa = 4.57TKK203 pKa = 8.8EE204 pKa = 3.95HH205 pKa = 6.25YY206 pKa = 10.44DD207 pKa = 3.35IIKK210 pKa = 10.21NYY212 pKa = 10.5YY213 pKa = 8.0YY214 pKa = 11.13YY215 pKa = 11.21KK216 pKa = 10.58FYY218 pKa = 11.18NEE220 pKa = 4.7KK221 pKa = 10.57LLSCLKK227 pKa = 10.8CIDD230 pKa = 3.56NLIYY234 pKa = 10.36ILQKK238 pKa = 10.54SGLNEE243 pKa = 4.24TYY245 pKa = 10.66CNEE248 pKa = 3.88IQLDD252 pKa = 4.2NINSIDD258 pKa = 3.85EE259 pKa = 4.34FEE261 pKa = 5.44LYY263 pKa = 10.99LFDD266 pKa = 4.9FVKK269 pKa = 10.78NNHH272 pKa = 6.12GSTEE276 pKa = 4.48FILCNINKK284 pKa = 9.89LYY286 pKa = 10.79KK287 pKa = 10.28DD288 pKa = 3.16IYY290 pKa = 7.77NTYY293 pKa = 10.13RR294 pKa = 11.84LIHH297 pKa = 6.45KK298 pKa = 9.79EE299 pKa = 3.88YY300 pKa = 11.01DD301 pKa = 3.26FLDD304 pKa = 4.39YY305 pKa = 10.52IPKK308 pKa = 8.96TFYY311 pKa = 10.17PQMMGFKK318 pKa = 9.85LAKK321 pKa = 9.21QNIVEE326 pKa = 4.48TCSGTFIKK334 pKa = 9.83IDD336 pKa = 3.46NKK338 pKa = 10.43LCPISKK344 pKa = 10.26CFFNKK349 pKa = 10.32LYY351 pKa = 8.8KK352 pKa = 9.28THH354 pKa = 6.79LRR356 pKa = 11.84SCPSYY361 pKa = 11.09SRR363 pKa = 11.84YY364 pKa = 10.58NYY366 pKa = 10.68DD367 pKa = 3.93GINYY371 pKa = 9.36LCRR374 pKa = 11.84TIQNRR379 pKa = 11.84IDD381 pKa = 3.25ILAAKK386 pKa = 9.97ANLYY390 pKa = 11.09CNDD393 pKa = 3.33KK394 pKa = 10.7NICKK398 pKa = 9.99LYY400 pKa = 10.94SMYY403 pKa = 11.12KK404 pKa = 9.88LVYY407 pKa = 7.55EE408 pKa = 4.53HH409 pKa = 7.29RR410 pKa = 11.84LIPISDD416 pKa = 3.7FNKK419 pKa = 9.44PLEE422 pKa = 3.95MLKK425 pKa = 10.38DD426 pKa = 3.58YY427 pKa = 10.95KK428 pKa = 11.21NYY430 pKa = 9.89MEE432 pKa = 5.15LEE434 pKa = 4.47SIFSFRR440 pKa = 11.84RR441 pKa = 11.84TFGIEE446 pKa = 4.01LNLDD450 pKa = 3.61NYY452 pKa = 9.9EE453 pKa = 4.08KK454 pKa = 10.87AIMHH458 pKa = 5.98PQFYY462 pKa = 9.35PYY464 pKa = 10.52RR465 pKa = 11.84SIFTSISNVIEE476 pKa = 4.06KK477 pKa = 10.68SEE479 pKa = 4.24IADD482 pKa = 3.64KK483 pKa = 9.27QHH485 pKa = 6.76KK486 pKa = 9.64EE487 pKa = 3.71EE488 pKa = 4.09NFEE491 pKa = 4.27HH492 pKa = 6.62KK493 pKa = 10.58KK494 pKa = 10.05HH495 pKa = 5.69VKK497 pKa = 10.03KK498 pKa = 10.77LLNSYY503 pKa = 9.8RR504 pKa = 11.84YY505 pKa = 9.44EE506 pKa = 4.14YY507 pKa = 11.42
Molecular weight: 60.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1563 |
92 |
583 |
390.8 |
44.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.566 ± 1.707 | 1.536 ± 0.718 |
4.798 ± 0.834 | 5.31 ± 1.061 |
5.054 ± 0.692 | 4.607 ± 0.549 |
1.536 ± 0.481 | 7.102 ± 0.959 |
7.422 ± 1.179 | 8.893 ± 0.77 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.791 ± 0.209 | 9.341 ± 0.517 |
3.263 ± 0.783 | 4.031 ± 1.181 |
3.263 ± 0.231 | 9.277 ± 0.971 |
6.206 ± 0.795 | 4.287 ± 0.836 |
0.512 ± 0.115 | 6.206 ± 1.884 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
