
Inachis io cypovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Cypovirus; Cypovirus 2
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
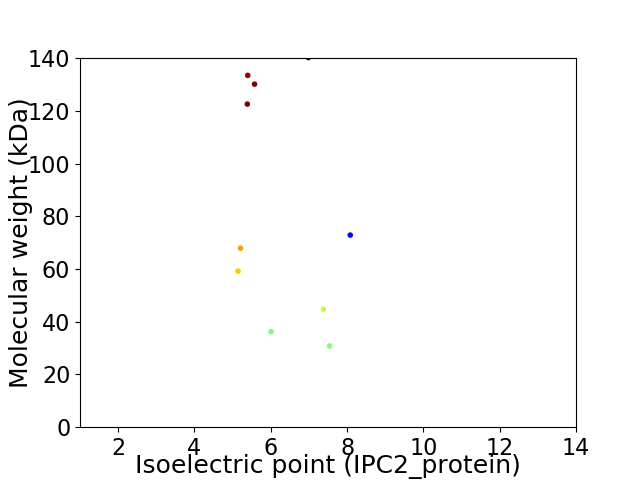
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W6EJ67|W6EJ67_9REOV VP5 OS=Inachis io cypovirus 2 OX=1382295 PE=4 SV=1
MM1 pKa = 8.0AYY3 pKa = 8.99PAQGYY8 pKa = 9.09VLDD11 pKa = 3.76EE12 pKa = 4.34RR13 pKa = 11.84FNFEE17 pKa = 3.91YY18 pKa = 10.34AIKK21 pKa = 10.42LLRR24 pKa = 11.84EE25 pKa = 3.54ADD27 pKa = 3.44INGEE31 pKa = 4.26TKK33 pKa = 10.76DD34 pKa = 3.62KK35 pKa = 11.41VSVAEE40 pKa = 4.37LRR42 pKa = 11.84DD43 pKa = 4.31FIIKK47 pKa = 9.43EE48 pKa = 4.0AQDD51 pKa = 3.12EE52 pKa = 4.32QFAVRR57 pKa = 11.84EE58 pKa = 4.17RR59 pKa = 11.84PLPPAQVRR67 pKa = 11.84GISARR72 pKa = 11.84VTVCPINSILCQLFCGDD89 pKa = 3.61GFYY92 pKa = 10.94AQSFIKK98 pKa = 10.53LMAIGQMYY106 pKa = 10.32ANQITAEE113 pKa = 4.05AFTVEE118 pKa = 3.99EE119 pKa = 4.24GQFYY123 pKa = 10.81VNEE126 pKa = 4.41GGIGPDD132 pKa = 3.02STKK135 pKa = 10.72YY136 pKa = 8.47PTAHH140 pKa = 6.36VAPLMEE146 pKa = 4.76AFRR149 pKa = 11.84RR150 pKa = 11.84CANAMDD156 pKa = 4.26STIDD160 pKa = 3.3PHH162 pKa = 6.69GTIQQLYY169 pKa = 9.69ALMEE173 pKa = 4.41EE174 pKa = 4.5TTHH177 pKa = 5.8MEE179 pKa = 4.08TWFMDD184 pKa = 3.7PSVITPRR191 pKa = 11.84QITSSIQNWNLFAAFVDD208 pKa = 4.03MSEE211 pKa = 4.36PNPVFPVATWQKK223 pKa = 10.97LLDD226 pKa = 3.53ILRR229 pKa = 11.84AVVLTFITSVSSTAVFDD246 pKa = 4.19EE247 pKa = 5.09GPPLQISFLLPSYY260 pKa = 10.75QLTNPRR266 pKa = 11.84LVNFTRR272 pKa = 11.84SYY274 pKa = 10.9QNLRR278 pKa = 11.84QQYY281 pKa = 9.2ISWILRR287 pKa = 11.84TVFIPFLEE295 pKa = 3.97RR296 pKa = 11.84LRR298 pKa = 11.84RR299 pKa = 11.84FIVGNNARR307 pKa = 11.84IARR310 pKa = 11.84TTFAGEE316 pKa = 3.93KK317 pKa = 9.74KK318 pKa = 10.31VAIPYY323 pKa = 10.25FGDD326 pKa = 3.45DD327 pKa = 4.96RR328 pKa = 11.84EE329 pKa = 4.38MKK331 pKa = 9.69TNNTIQVVDD340 pKa = 3.2KK341 pKa = 10.71RR342 pKa = 11.84GRR344 pKa = 11.84ANYY347 pKa = 9.51DD348 pKa = 2.96AYY350 pKa = 10.07KK351 pKa = 9.42YY352 pKa = 9.05TLEE355 pKa = 3.78NHH357 pKa = 6.61LLFGEE362 pKa = 4.39DD363 pKa = 2.93GRR365 pKa = 11.84RR366 pKa = 11.84PRR368 pKa = 11.84MFTKK372 pKa = 10.32TLVQNDD378 pKa = 4.17DD379 pKa = 3.41LSTFHH384 pKa = 6.46VQIFNSFTDD393 pKa = 4.3TIYY396 pKa = 9.67GTNKK400 pKa = 9.98NVTAAYY406 pKa = 8.25DD407 pKa = 3.53TMSALVAKK415 pKa = 7.24YY416 pKa = 8.85TPINYY421 pKa = 7.54NTLIVEE427 pKa = 4.37SLDD430 pKa = 3.44DD431 pKa = 3.83YY432 pKa = 12.01NRR434 pKa = 11.84INLKK438 pKa = 9.47TGDD441 pKa = 3.55ISQVAVWFLTEE452 pKa = 3.37YY453 pKa = 10.4SRR455 pKa = 11.84PYY457 pKa = 10.65VYY459 pKa = 10.52FGVASLYY466 pKa = 11.19LLDD469 pKa = 4.47DD470 pKa = 4.37TIIAYY475 pKa = 7.47TVHH478 pKa = 6.19TNVGRR483 pKa = 11.84YY484 pKa = 9.0HH485 pKa = 6.68EE486 pKa = 4.67GEE488 pKa = 3.8KK489 pKa = 10.06TFAFEE494 pKa = 4.13EE495 pKa = 4.7CTAMHH500 pKa = 7.05MFNILYY506 pKa = 10.03SLQVCLDD513 pKa = 3.55PEE515 pKa = 4.31AFLTNGEE522 pKa = 4.19MDD524 pKa = 3.17KK525 pKa = 11.25HH526 pKa = 6.68EE527 pKa = 4.29YY528 pKa = 8.7TLDD531 pKa = 3.39YY532 pKa = 10.6RR533 pKa = 11.84VWRR536 pKa = 11.84EE537 pKa = 3.69ALSQEE542 pKa = 5.47LKK544 pKa = 9.01TCWRR548 pKa = 11.84PLCDD552 pKa = 3.06HH553 pKa = 6.11YY554 pKa = 11.72ARR556 pKa = 11.84RR557 pKa = 11.84NVQDD561 pKa = 3.59TPTVAMAEE569 pKa = 4.35MKK571 pKa = 10.02EE572 pKa = 4.05LMGAYY577 pKa = 9.05PYY579 pKa = 9.37TAIYY583 pKa = 9.94KK584 pKa = 10.41FEE586 pKa = 4.64DD587 pKa = 3.24VHH589 pKa = 5.82TAA591 pKa = 2.97
MM1 pKa = 8.0AYY3 pKa = 8.99PAQGYY8 pKa = 9.09VLDD11 pKa = 3.76EE12 pKa = 4.34RR13 pKa = 11.84FNFEE17 pKa = 3.91YY18 pKa = 10.34AIKK21 pKa = 10.42LLRR24 pKa = 11.84EE25 pKa = 3.54ADD27 pKa = 3.44INGEE31 pKa = 4.26TKK33 pKa = 10.76DD34 pKa = 3.62KK35 pKa = 11.41VSVAEE40 pKa = 4.37LRR42 pKa = 11.84DD43 pKa = 4.31FIIKK47 pKa = 9.43EE48 pKa = 4.0AQDD51 pKa = 3.12EE52 pKa = 4.32QFAVRR57 pKa = 11.84EE58 pKa = 4.17RR59 pKa = 11.84PLPPAQVRR67 pKa = 11.84GISARR72 pKa = 11.84VTVCPINSILCQLFCGDD89 pKa = 3.61GFYY92 pKa = 10.94AQSFIKK98 pKa = 10.53LMAIGQMYY106 pKa = 10.32ANQITAEE113 pKa = 4.05AFTVEE118 pKa = 3.99EE119 pKa = 4.24GQFYY123 pKa = 10.81VNEE126 pKa = 4.41GGIGPDD132 pKa = 3.02STKK135 pKa = 10.72YY136 pKa = 8.47PTAHH140 pKa = 6.36VAPLMEE146 pKa = 4.76AFRR149 pKa = 11.84RR150 pKa = 11.84CANAMDD156 pKa = 4.26STIDD160 pKa = 3.3PHH162 pKa = 6.69GTIQQLYY169 pKa = 9.69ALMEE173 pKa = 4.41EE174 pKa = 4.5TTHH177 pKa = 5.8MEE179 pKa = 4.08TWFMDD184 pKa = 3.7PSVITPRR191 pKa = 11.84QITSSIQNWNLFAAFVDD208 pKa = 4.03MSEE211 pKa = 4.36PNPVFPVATWQKK223 pKa = 10.97LLDD226 pKa = 3.53ILRR229 pKa = 11.84AVVLTFITSVSSTAVFDD246 pKa = 4.19EE247 pKa = 5.09GPPLQISFLLPSYY260 pKa = 10.75QLTNPRR266 pKa = 11.84LVNFTRR272 pKa = 11.84SYY274 pKa = 10.9QNLRR278 pKa = 11.84QQYY281 pKa = 9.2ISWILRR287 pKa = 11.84TVFIPFLEE295 pKa = 3.97RR296 pKa = 11.84LRR298 pKa = 11.84RR299 pKa = 11.84FIVGNNARR307 pKa = 11.84IARR310 pKa = 11.84TTFAGEE316 pKa = 3.93KK317 pKa = 9.74KK318 pKa = 10.31VAIPYY323 pKa = 10.25FGDD326 pKa = 3.45DD327 pKa = 4.96RR328 pKa = 11.84EE329 pKa = 4.38MKK331 pKa = 9.69TNNTIQVVDD340 pKa = 3.2KK341 pKa = 10.71RR342 pKa = 11.84GRR344 pKa = 11.84ANYY347 pKa = 9.51DD348 pKa = 2.96AYY350 pKa = 10.07KK351 pKa = 9.42YY352 pKa = 9.05TLEE355 pKa = 3.78NHH357 pKa = 6.61LLFGEE362 pKa = 4.39DD363 pKa = 2.93GRR365 pKa = 11.84RR366 pKa = 11.84PRR368 pKa = 11.84MFTKK372 pKa = 10.32TLVQNDD378 pKa = 4.17DD379 pKa = 3.41LSTFHH384 pKa = 6.46VQIFNSFTDD393 pKa = 4.3TIYY396 pKa = 9.67GTNKK400 pKa = 9.98NVTAAYY406 pKa = 8.25DD407 pKa = 3.53TMSALVAKK415 pKa = 7.24YY416 pKa = 8.85TPINYY421 pKa = 7.54NTLIVEE427 pKa = 4.37SLDD430 pKa = 3.44DD431 pKa = 3.83YY432 pKa = 12.01NRR434 pKa = 11.84INLKK438 pKa = 9.47TGDD441 pKa = 3.55ISQVAVWFLTEE452 pKa = 3.37YY453 pKa = 10.4SRR455 pKa = 11.84PYY457 pKa = 10.65VYY459 pKa = 10.52FGVASLYY466 pKa = 11.19LLDD469 pKa = 4.47DD470 pKa = 4.37TIIAYY475 pKa = 7.47TVHH478 pKa = 6.19TNVGRR483 pKa = 11.84YY484 pKa = 9.0HH485 pKa = 6.68EE486 pKa = 4.67GEE488 pKa = 3.8KK489 pKa = 10.06TFAFEE494 pKa = 4.13EE495 pKa = 4.7CTAMHH500 pKa = 7.05MFNILYY506 pKa = 10.03SLQVCLDD513 pKa = 3.55PEE515 pKa = 4.31AFLTNGEE522 pKa = 4.19MDD524 pKa = 3.17KK525 pKa = 11.25HH526 pKa = 6.68EE527 pKa = 4.29YY528 pKa = 8.7TLDD531 pKa = 3.39YY532 pKa = 10.6RR533 pKa = 11.84VWRR536 pKa = 11.84EE537 pKa = 3.69ALSQEE542 pKa = 5.47LKK544 pKa = 9.01TCWRR548 pKa = 11.84PLCDD552 pKa = 3.06HH553 pKa = 6.11YY554 pKa = 11.72ARR556 pKa = 11.84RR557 pKa = 11.84NVQDD561 pKa = 3.59TPTVAMAEE569 pKa = 4.35MKK571 pKa = 10.02EE572 pKa = 4.05LMGAYY577 pKa = 9.05PYY579 pKa = 9.37TAIYY583 pKa = 9.94KK584 pKa = 10.41FEE586 pKa = 4.64DD587 pKa = 3.24VHH589 pKa = 5.82TAA591 pKa = 2.97
Molecular weight: 67.92 kDa
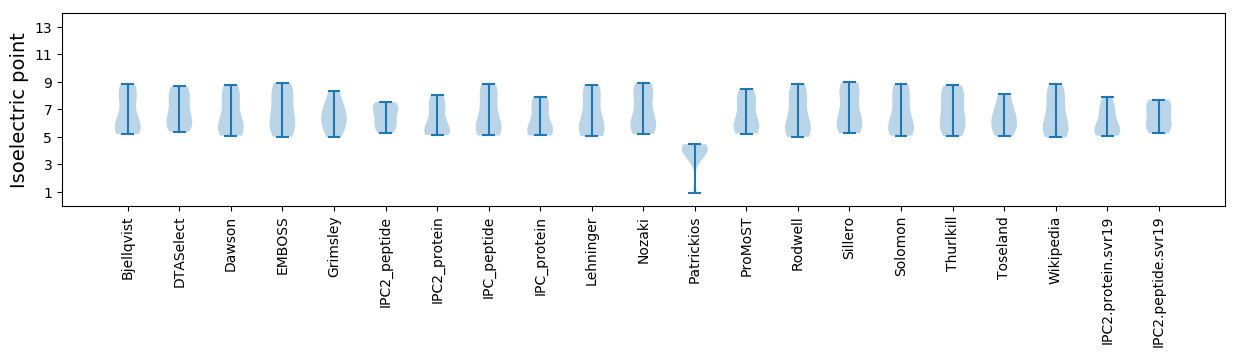
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6EJ73|W6EJ73_9REOV Polyhedrin OS=Inachis io cypovirus 2 OX=1382295 PE=4 SV=1
MM1 pKa = 7.44EE2 pKa = 5.88LVPLTASNAEE12 pKa = 4.25LMLKK16 pKa = 8.49TYY18 pKa = 10.41CLAVRR23 pKa = 11.84PRR25 pKa = 11.84GKK27 pKa = 10.13KK28 pKa = 8.67GQPNKK33 pKa = 10.2VSTLKK38 pKa = 10.57IPNSYY43 pKa = 10.68YY44 pKa = 10.71KK45 pKa = 10.68GIEE48 pKa = 3.98KK49 pKa = 10.18PSYY52 pKa = 8.68TKK54 pKa = 10.54NVPQLPIQVLSIEE67 pKa = 4.5TQLKK71 pKa = 9.19VHH73 pKa = 6.91DD74 pKa = 3.81KK75 pKa = 10.36LRR77 pKa = 11.84RR78 pKa = 11.84IFEE81 pKa = 4.39RR82 pKa = 11.84NKK84 pKa = 10.3KK85 pKa = 10.07SYY87 pKa = 10.63EE88 pKa = 3.9NPRR91 pKa = 11.84LIARR95 pKa = 11.84LLTFGLTPKK104 pKa = 10.78GKK106 pKa = 9.13IDD108 pKa = 3.45MKK110 pKa = 11.28AFIDD114 pKa = 4.9SKK116 pKa = 11.15DD117 pKa = 3.15KK118 pKa = 11.01RR119 pKa = 11.84QNDD122 pKa = 3.63NKK124 pKa = 10.89DD125 pKa = 3.39VGPGRR130 pKa = 11.84SKK132 pKa = 10.74KK133 pKa = 9.9PKK135 pKa = 10.45LFGADD140 pKa = 3.13KK141 pKa = 11.06QDD143 pKa = 3.79FKK145 pKa = 11.64VKK147 pKa = 10.27KK148 pKa = 9.42HH149 pKa = 6.45DD150 pKa = 3.71VEE152 pKa = 4.5QSINDD157 pKa = 3.78DD158 pKa = 3.38FDD160 pKa = 4.72INAVHH165 pKa = 6.44SVDD168 pKa = 3.47GVRR171 pKa = 11.84DD172 pKa = 3.81LLEE175 pKa = 5.91LDD177 pKa = 4.59DD178 pKa = 5.33LRR180 pKa = 11.84GPLGITVVYY189 pKa = 8.73MINHH193 pKa = 6.7LFYY196 pKa = 10.52RR197 pKa = 11.84SKK199 pKa = 10.44IPAYY203 pKa = 7.27MAPLIPHH210 pKa = 6.62LFIRR214 pKa = 11.84KK215 pKa = 9.1INVNEE220 pKa = 3.94NMIPPLDD227 pKa = 3.59EE228 pKa = 5.89RR229 pKa = 11.84YY230 pKa = 9.28LQKK233 pKa = 10.37IYY235 pKa = 11.0LPVYY239 pKa = 7.49TASNILSCSIEE250 pKa = 4.22SSVEE254 pKa = 3.78SCIIAIDD261 pKa = 3.97KK262 pKa = 10.42QLSLADD268 pKa = 3.32KK269 pKa = 10.81QYY271 pKa = 11.01FDD273 pKa = 4.08KK274 pKa = 11.25HH275 pKa = 3.97FTNYY279 pKa = 9.94RR280 pKa = 11.84MIEE283 pKa = 4.1PGCALLMYY291 pKa = 10.54RR292 pKa = 11.84NYY294 pKa = 10.61TNNEE298 pKa = 4.08LCWIMSGLMLIEE310 pKa = 3.85AHH312 pKa = 6.89KK313 pKa = 10.46MIGKK317 pKa = 6.32TAKK320 pKa = 10.06EE321 pKa = 4.26SFNTLYY327 pKa = 9.42THH329 pKa = 7.63AYY331 pKa = 6.13EE332 pKa = 4.33TTVDD336 pKa = 3.38RR337 pKa = 11.84RR338 pKa = 11.84IVGGMKK344 pKa = 10.17VFIEE348 pKa = 3.97PTKK351 pKa = 10.53NYY353 pKa = 9.29VLEE356 pKa = 4.2RR357 pKa = 11.84VFPSGNPNVNPIILLRR373 pKa = 11.84TRR375 pKa = 11.84EE376 pKa = 3.7FFEE379 pKa = 4.05RR380 pKa = 11.84VKK382 pKa = 10.88LRR384 pKa = 11.84EE385 pKa = 3.92EE386 pKa = 4.49GKK388 pKa = 8.58FTGMNIYY395 pKa = 10.54GIIANKK401 pKa = 8.62GTGKK405 pKa = 8.44PTLSRR410 pKa = 11.84MLVEE414 pKa = 3.94RR415 pKa = 11.84LEE417 pKa = 4.19NKK419 pKa = 10.5GIGLSVIDD427 pKa = 3.53SDD429 pKa = 5.16EE430 pKa = 4.12YY431 pKa = 11.46GRR433 pKa = 11.84WLTMLLEE440 pKa = 4.41SSLINEE446 pKa = 4.86DD447 pKa = 4.12FSITEE452 pKa = 4.39PLDD455 pKa = 3.19EE456 pKa = 4.86YY457 pKa = 11.66YY458 pKa = 10.14PGRR461 pKa = 11.84PKK463 pKa = 10.66FKK465 pKa = 10.31LSEE468 pKa = 3.72RR469 pKa = 11.84SFFNIIMCMLLKK481 pKa = 10.32KK482 pKa = 10.16HH483 pKa = 6.62SITSRR488 pKa = 11.84DD489 pKa = 3.66KK490 pKa = 10.87LTLSYY495 pKa = 10.14YY496 pKa = 6.43TTRR499 pKa = 11.84CEE501 pKa = 4.93EE502 pKa = 3.89LFNDD506 pKa = 3.7YY507 pKa = 10.89KK508 pKa = 11.17EE509 pKa = 4.24YY510 pKa = 10.15LQKK513 pKa = 10.83VYY515 pKa = 9.75STEE518 pKa = 4.17SYY520 pKa = 11.55SMQAYY525 pKa = 9.42YY526 pKa = 10.6DD527 pKa = 3.57HH528 pKa = 7.67RR529 pKa = 11.84LPQCEE534 pKa = 4.15HH535 pKa = 6.43KK536 pKa = 10.96NIIVEE541 pKa = 4.2CHH543 pKa = 4.32TTLDD547 pKa = 4.02NVRR550 pKa = 11.84STPSDD555 pKa = 3.37LFVRR559 pKa = 11.84LGAFFDD565 pKa = 4.04PVVTITLIRR574 pKa = 11.84PSAKK578 pKa = 8.66EE579 pKa = 3.61TAKK582 pKa = 10.27TGLFHH587 pKa = 8.11LGEE590 pKa = 4.32LALYY594 pKa = 9.73YY595 pKa = 10.34YY596 pKa = 10.77YY597 pKa = 9.93EE598 pKa = 4.34TATVPDD604 pKa = 3.64VALVYY609 pKa = 9.86PSNVLYY615 pKa = 10.85ALDD618 pKa = 4.46LEE620 pKa = 4.9LGKK623 pKa = 8.78TPDD626 pKa = 3.13IVQVSSEE633 pKa = 3.88
MM1 pKa = 7.44EE2 pKa = 5.88LVPLTASNAEE12 pKa = 4.25LMLKK16 pKa = 8.49TYY18 pKa = 10.41CLAVRR23 pKa = 11.84PRR25 pKa = 11.84GKK27 pKa = 10.13KK28 pKa = 8.67GQPNKK33 pKa = 10.2VSTLKK38 pKa = 10.57IPNSYY43 pKa = 10.68YY44 pKa = 10.71KK45 pKa = 10.68GIEE48 pKa = 3.98KK49 pKa = 10.18PSYY52 pKa = 8.68TKK54 pKa = 10.54NVPQLPIQVLSIEE67 pKa = 4.5TQLKK71 pKa = 9.19VHH73 pKa = 6.91DD74 pKa = 3.81KK75 pKa = 10.36LRR77 pKa = 11.84RR78 pKa = 11.84IFEE81 pKa = 4.39RR82 pKa = 11.84NKK84 pKa = 10.3KK85 pKa = 10.07SYY87 pKa = 10.63EE88 pKa = 3.9NPRR91 pKa = 11.84LIARR95 pKa = 11.84LLTFGLTPKK104 pKa = 10.78GKK106 pKa = 9.13IDD108 pKa = 3.45MKK110 pKa = 11.28AFIDD114 pKa = 4.9SKK116 pKa = 11.15DD117 pKa = 3.15KK118 pKa = 11.01RR119 pKa = 11.84QNDD122 pKa = 3.63NKK124 pKa = 10.89DD125 pKa = 3.39VGPGRR130 pKa = 11.84SKK132 pKa = 10.74KK133 pKa = 9.9PKK135 pKa = 10.45LFGADD140 pKa = 3.13KK141 pKa = 11.06QDD143 pKa = 3.79FKK145 pKa = 11.64VKK147 pKa = 10.27KK148 pKa = 9.42HH149 pKa = 6.45DD150 pKa = 3.71VEE152 pKa = 4.5QSINDD157 pKa = 3.78DD158 pKa = 3.38FDD160 pKa = 4.72INAVHH165 pKa = 6.44SVDD168 pKa = 3.47GVRR171 pKa = 11.84DD172 pKa = 3.81LLEE175 pKa = 5.91LDD177 pKa = 4.59DD178 pKa = 5.33LRR180 pKa = 11.84GPLGITVVYY189 pKa = 8.73MINHH193 pKa = 6.7LFYY196 pKa = 10.52RR197 pKa = 11.84SKK199 pKa = 10.44IPAYY203 pKa = 7.27MAPLIPHH210 pKa = 6.62LFIRR214 pKa = 11.84KK215 pKa = 9.1INVNEE220 pKa = 3.94NMIPPLDD227 pKa = 3.59EE228 pKa = 5.89RR229 pKa = 11.84YY230 pKa = 9.28LQKK233 pKa = 10.37IYY235 pKa = 11.0LPVYY239 pKa = 7.49TASNILSCSIEE250 pKa = 4.22SSVEE254 pKa = 3.78SCIIAIDD261 pKa = 3.97KK262 pKa = 10.42QLSLADD268 pKa = 3.32KK269 pKa = 10.81QYY271 pKa = 11.01FDD273 pKa = 4.08KK274 pKa = 11.25HH275 pKa = 3.97FTNYY279 pKa = 9.94RR280 pKa = 11.84MIEE283 pKa = 4.1PGCALLMYY291 pKa = 10.54RR292 pKa = 11.84NYY294 pKa = 10.61TNNEE298 pKa = 4.08LCWIMSGLMLIEE310 pKa = 3.85AHH312 pKa = 6.89KK313 pKa = 10.46MIGKK317 pKa = 6.32TAKK320 pKa = 10.06EE321 pKa = 4.26SFNTLYY327 pKa = 9.42THH329 pKa = 7.63AYY331 pKa = 6.13EE332 pKa = 4.33TTVDD336 pKa = 3.38RR337 pKa = 11.84RR338 pKa = 11.84IVGGMKK344 pKa = 10.17VFIEE348 pKa = 3.97PTKK351 pKa = 10.53NYY353 pKa = 9.29VLEE356 pKa = 4.2RR357 pKa = 11.84VFPSGNPNVNPIILLRR373 pKa = 11.84TRR375 pKa = 11.84EE376 pKa = 3.7FFEE379 pKa = 4.05RR380 pKa = 11.84VKK382 pKa = 10.88LRR384 pKa = 11.84EE385 pKa = 3.92EE386 pKa = 4.49GKK388 pKa = 8.58FTGMNIYY395 pKa = 10.54GIIANKK401 pKa = 8.62GTGKK405 pKa = 8.44PTLSRR410 pKa = 11.84MLVEE414 pKa = 3.94RR415 pKa = 11.84LEE417 pKa = 4.19NKK419 pKa = 10.5GIGLSVIDD427 pKa = 3.53SDD429 pKa = 5.16EE430 pKa = 4.12YY431 pKa = 11.46GRR433 pKa = 11.84WLTMLLEE440 pKa = 4.41SSLINEE446 pKa = 4.86DD447 pKa = 4.12FSITEE452 pKa = 4.39PLDD455 pKa = 3.19EE456 pKa = 4.86YY457 pKa = 11.66YY458 pKa = 10.14PGRR461 pKa = 11.84PKK463 pKa = 10.66FKK465 pKa = 10.31LSEE468 pKa = 3.72RR469 pKa = 11.84SFFNIIMCMLLKK481 pKa = 10.32KK482 pKa = 10.16HH483 pKa = 6.62SITSRR488 pKa = 11.84DD489 pKa = 3.66KK490 pKa = 10.87LTLSYY495 pKa = 10.14YY496 pKa = 6.43TTRR499 pKa = 11.84CEE501 pKa = 4.93EE502 pKa = 3.89LFNDD506 pKa = 3.7YY507 pKa = 10.89KK508 pKa = 11.17EE509 pKa = 4.24YY510 pKa = 10.15LQKK513 pKa = 10.83VYY515 pKa = 9.75STEE518 pKa = 4.17SYY520 pKa = 11.55SMQAYY525 pKa = 9.42YY526 pKa = 10.6DD527 pKa = 3.57HH528 pKa = 7.67RR529 pKa = 11.84LPQCEE534 pKa = 4.15HH535 pKa = 6.43KK536 pKa = 10.96NIIVEE541 pKa = 4.2CHH543 pKa = 4.32TTLDD547 pKa = 4.02NVRR550 pKa = 11.84STPSDD555 pKa = 3.37LFVRR559 pKa = 11.84LGAFFDD565 pKa = 4.04PVVTITLIRR574 pKa = 11.84PSAKK578 pKa = 8.66EE579 pKa = 3.61TAKK582 pKa = 10.27TGLFHH587 pKa = 8.11LGEE590 pKa = 4.32LALYY594 pKa = 9.73YY595 pKa = 10.34YY596 pKa = 10.77YY597 pKa = 9.93EE598 pKa = 4.34TATVPDD604 pKa = 3.64VALVYY609 pKa = 9.86PSNVLYY615 pKa = 10.85ALDD618 pKa = 4.46LEE620 pKa = 4.9LGKK623 pKa = 8.78TPDD626 pKa = 3.13IVQVSSEE633 pKa = 3.88
Molecular weight: 72.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7389 |
274 |
1225 |
738.9 |
83.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.968 ± 0.327 | 1.164 ± 0.148 |
6.009 ± 0.247 | 5.305 ± 0.277 |
4.629 ± 0.318 | 4.98 ± 0.192 |
2.477 ± 0.188 | 7.484 ± 0.297 |
4.913 ± 0.669 | 8.932 ± 0.485 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.199 | 6.401 ± 0.37 |
4.453 ± 0.256 | 3.451 ± 0.194 |
5.265 ± 0.234 | 7.498 ± 0.631 |
7.065 ± 0.403 | 5.968 ± 0.282 |
0.717 ± 0.145 | 4.656 ± 0.473 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
